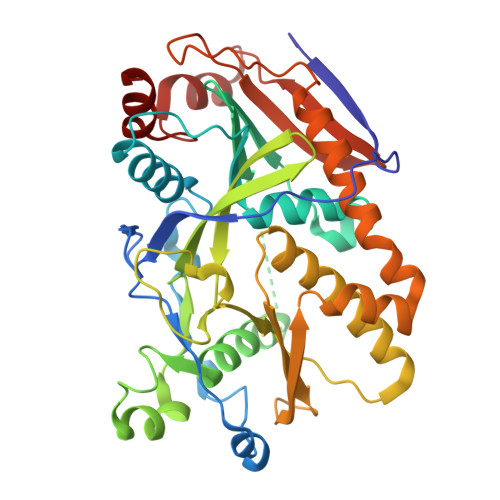
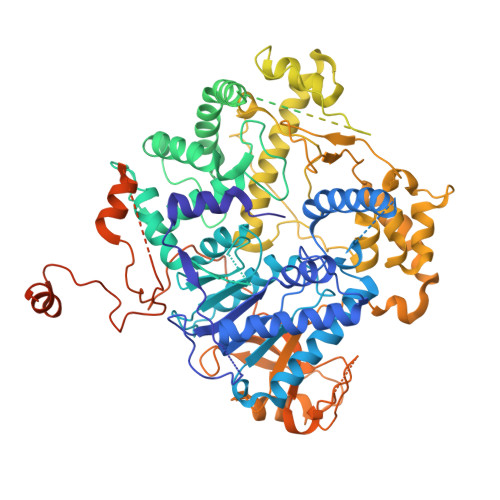
Assembly of the Xrn2/Rat1-Rai1-Rtt103 termination complexes in mesophilic and thermophilic organisms.
Dikunova, A., Noskova, N., Overbeck, J.H., Polak, M., Stelzig, D., Zapletal, D., Kubicek, K., Novacek, J., Sprangers, R., Stefl, R.(2024) Structure
- PubMed: 39657659
- DOI: https://doi.org/10.1016/j.str.2024.11.010
- Primary Citation of Related Structures:
8Q6V, 9EXS, 9FMS - PubMed Abstract:
The 5'-3' exoribonuclease Xrn2, known as Rat1 in yeasts, terminates mRNA transcription by RNA polymerase II (RNAPII). In the torpedo model of termination, the activity of Xrn2/Rat1 is enhanced by Rai1, which is recruited to the termination site by Rtt103, an adaptor protein binding to the RNAPII C-terminal domain (CTD). The overall architecture of the Xrn2/Rat1-Rai1-Rtt103 complex remains unknown. We combined structural biology methods to characterize the torpedo complex from Saccharomyces cerevisiae and Chaetomium thermophilum. Comparison of the structures from these organisms revealed a conserved protein core fold of the subunits, but significant variability in their interaction interfaces. We found that in the mesophile, Rtt103 utilizes an unstructured region to augment a Rai1 β-sheet, while in the thermophile Rtt103 binds to a C-terminal helix of Rai1 via its CTD-interacting domain with an α-helical fold. These different torpedo complex assemblies reflect adaptations to the environment and impact complex recruitment to RNAPII.
Organizational Affiliation:
CEITEC-Central European Institute of Technology, Masaryk University, Brno, Czechia; National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Brno, Czechia.