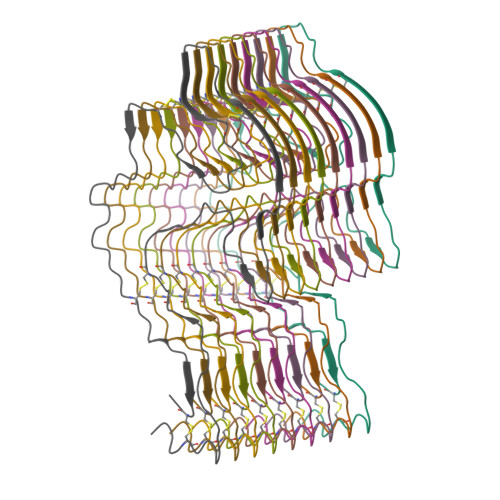
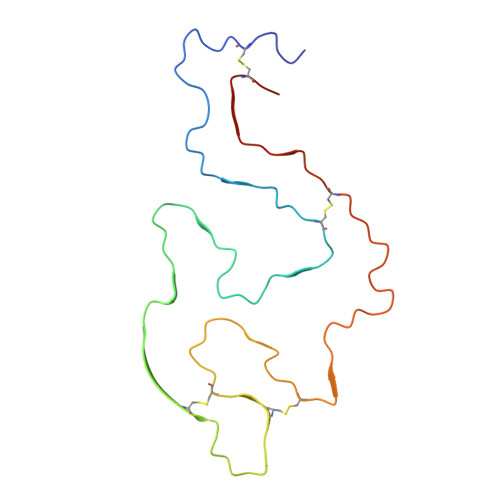
Cryo-EM structure of a lysozyme-derived amyloid fibril from hereditary amyloidosis.
Karimi-Farsijani, S., Sharma, K., Ugrina, M., Kuhn, L., Pfeiffer, P.B., Haupt, C., Wiese, S., Hegenbart, U., Schonland, S.O., Schwierz, N., Schmidt, M., Fandrich, M.(2024) Nat Commun 15: 9648-9648
- PubMed: 39511224
- DOI: https://doi.org/10.1038/s41467-024-54091-7
- Primary Citation of Related Structures:
8R4A - PubMed Abstract:
Systemic ALys amyloidosis is a debilitating protein misfolding disease that arises from the formation of amyloid fibrils from C-type lysozyme. We here present a 2.8 Å cryo-electron microscopy structure of an amyloid fibril, which was isolated from the abdominal fat tissue of a patient who expressed the D87G variant of human lysozyme. We find that the fibril possesses a stable core that is formed by all 130 residues of the fibril precursor protein. There are four disulfide bonds in each fibril protein that connect the same residues as in the globularly folded protein. As the conformation of lysozyme in the fibril is otherwise fundamentally different from native lysozyme, our data provide a structural rationale for the need of protein unfolding in the development of systemic ALys amyloidosis.
Organizational Affiliation:
Institute of Protein Biochemistry, Ulm University, Ulm, Germany. sara.karimi-farsijani@uni-ulm.de.