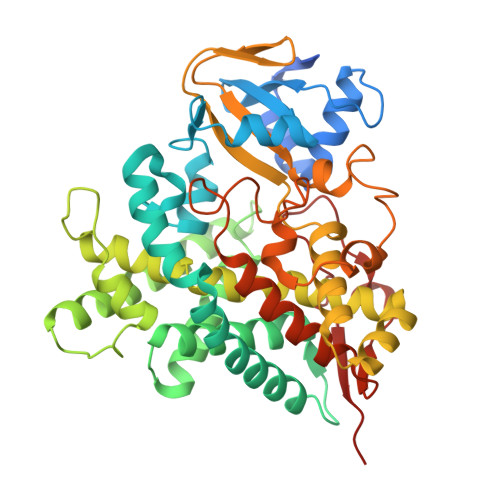
Coordinated conformational changes play a pivotal role in unsaturated fatty acid decarboxylation into renewable olefin
Generoso, W.C., Alvarenga, A.H.S., Simoes, I.T., Miyamoto, R.Y., Mandelli, F., Santos, C.A., Santos, C.R., Colombari, F.M., Morais, M.A.B., Fernandes, R., Persinoti, G.F., Murakami, M.T., Zanphorlin, L.M.To be published.