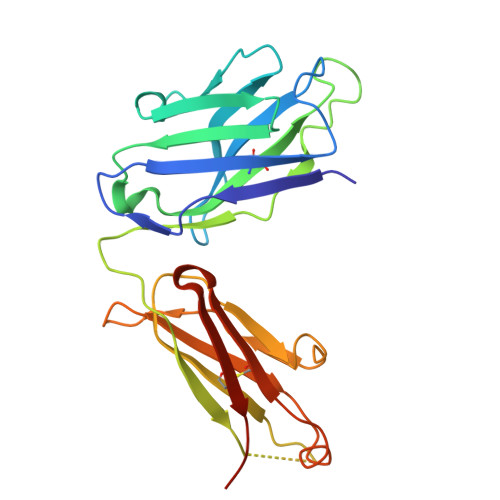
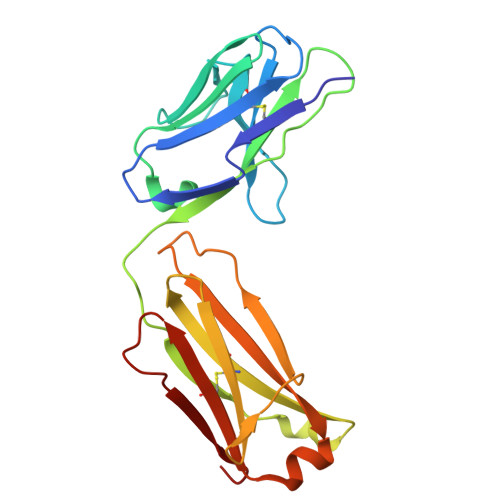
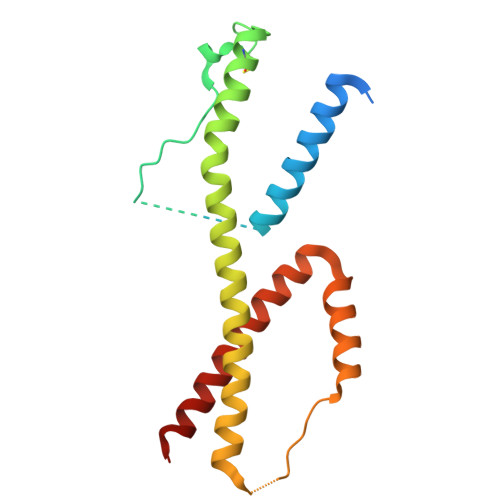
Structural basis of signaling complex inhibition by IL-6 domain-swapped dimers.
Yudenko, A., Bukhdruker, S., Shishkin, P., Rodin, S., Burtseva, A., Petrov, A., Pigareva, N., Sokolov, A., Zinovev, E., Eliseev, I., Remeeva, A., Marin, E., Mishin, A., Gordeliy, V., Gushchin, I., Ischenko, A., Borshchevskiy, V.(2025) Structure 33: 171-180.e5
- PubMed: 39566503
- DOI: https://doi.org/10.1016/j.str.2024.10.028
- Primary Citation of Related Structures:
8YWP, 8YWQ, 8YWR - PubMed Abstract:
Interleukin-6 (IL-6) is a multifaceted cytokine essential in many immune system processes and their regulation. It also plays a key role in hematopoiesis, and in triggering the acute phase reaction. IL-6 overproduction is critical in chronic inflammation associated with autoimmune diseases like rheumatoid arthritis and contributes to cytokine storms in COVID-19 patients. Over 20 years ago, researchers proposed that IL-6, which is typically monomeric, can also form dimers via a domain-swap mechanism, with indirect evidence supporting their existence. The physiological significance of IL-6 dimers was shown in B-cell chronic lymphocytic leukemia. However, no structures have been reported so far. Here, we present the crystal structure of an IL-6 domain-swapped dimer that computational approaches could not predict. The structure explains why the IL-6 dimer is antagonistic to the IL-6 monomer in signaling complex formation and provides insights for IL-6 targeted therapies.
Organizational Affiliation:
Research Center for Molecular Mechanisms of Aging and Age-Related Diseases, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region 141701, Russia.