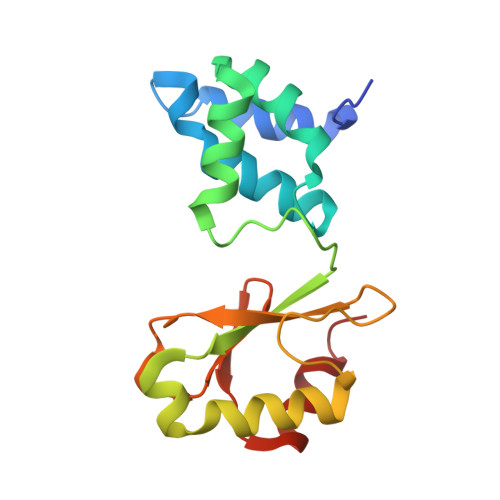
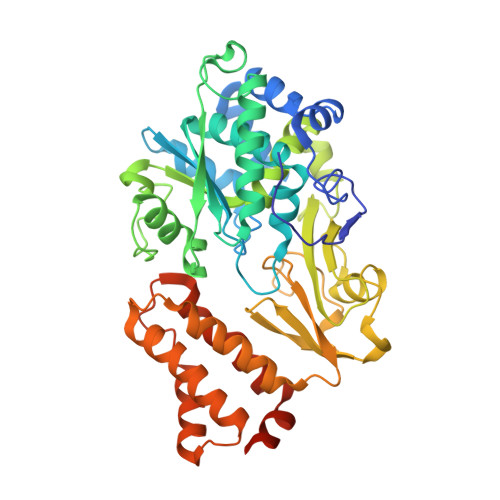
Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Goppert-Asadollahpour, S., Wohlwend, D., Friedrich, T.(2024) Biochim Biophys Acta Bioenerg 1865: 149491-149491
- PubMed: 38960077
- DOI: https://doi.org/10.1016/j.bbabio.2024.149491
- Primary Citation of Related Structures:
9FDJ, 9FDK, 9FDV, 9FE0, 9FE5, 9FE7, 9FE8, 9FEA, 9FIF, 9FIH, 9FII, 9FIJ, 9FIL - PubMed Abstract:
Energy converting NADH:ubiquinone oxidoreductase, complex I, is the first enzyme of respiratory chains in most eukaryotes and many bacteria. Mutations in genes encoding subunits of human complex I may lead to its dysfunction resulting in a diverse clinical pattern. The effect of mutations on the protein structure is not known. Here, we focus on mutations R88G, E246K, P252R and E377K that are found in subunit NDUFV1 comprising the NADH binding site of complex I. Homologous mutations were introduced into subunit NuoF of Aquifex aeolicus complex I and it was attempted to crystallize variants of the electron input module, NuoEF, with bound substrates in the oxidized and reduced state. The E377K variant did not form crystals most likely due to an improper protein assembly. The architecture of the NADH binding site is hardly affected by the other mutations indicating its unexpected structural robustness. The R88G, E246K and P252R mutations led to small local structural rearrangements that might be related to their pathogenicity. These minor structural changes involve substrate binding, product release and the putative formation of reactive oxygen species. The structural consequences of the mutations as obtained with the bacterial enzyme might thus help to contribute to the understanding of disease causing mutations.
- Albert-Ludwigs-Universität Freiburg, Institut für Biochemie, Albertstr. 21, D-79104 Freiburg, Germany.
Organizational Affiliation: