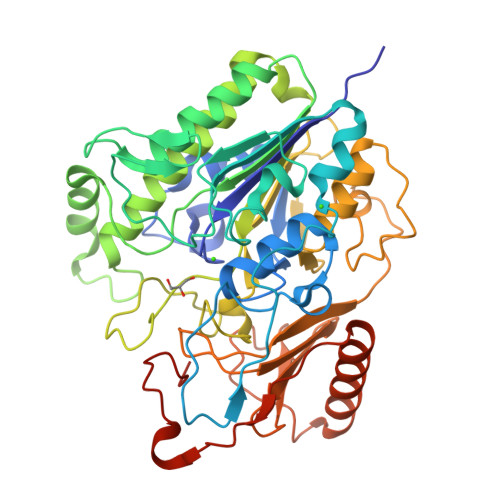
Identification and Characterization of a New Thermophilic kappa-Carrageenan Sulfatase.
Rhein-Knudsen, N., Reyes-Weiss, D.S., Klau, L.J., Jeudy, A., Roret, T., Stokke, R., Eijsink, V.G.H., Aachmann, F.L., Czjzek, M., Horn, S.J.(2025) J Agric Food Chem 73: 2044-2055
- PubMed: 39797788
- DOI: https://doi.org/10.1021/acs.jafc.4c09751
- Primary Citation of Related Structures:
9FO1 - PubMed Abstract:
Carrageenans are sulfated polysaccharides found in the cell wall of certain red seaweeds. They are widely used in the food industry for their gelling and stabilizing properties. In nature, carrageenans undergo enzymatic modification and degradation by marine organisms. Characterizing these enzymes is crucial for understanding carrageenan utilization and may eventually enable the development of targeted processes to modify carrageenans for industrial applications. In our study, we characterized a κ-carrageenan sulfatase, AMOR_S1_16A, belonging to the sulfatase S1_16 subfamily, which selectively desulfates the nonreducing end galactoses of κ-carrageenan oligomers in an exomode. Notably, AMOR_S1_16A represents the first κ-carrageenan sulfatase within the S1_16 subfamily and exhibits a novel enzymatic activity. This study provides further understanding of the substrate specificity and characteristics of the S1_16 subfamily. Moreover, this research highlights that many processes and enzymes remain to be discovered to fully understand carrageenan utilization pathways and to develop enzymatic processes for carrageenan modification and processing.
Organizational Affiliation:
Faculty of Chemistry, Biotechnology, and Food Science, NMBU Norwegian University of Life Sciences, P.O. Box 5003, 1432 Aas, Norway.