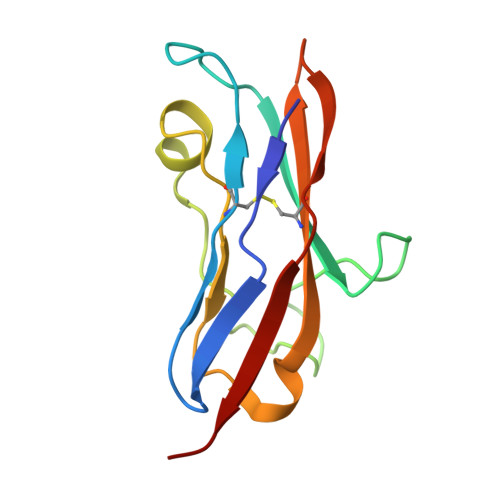
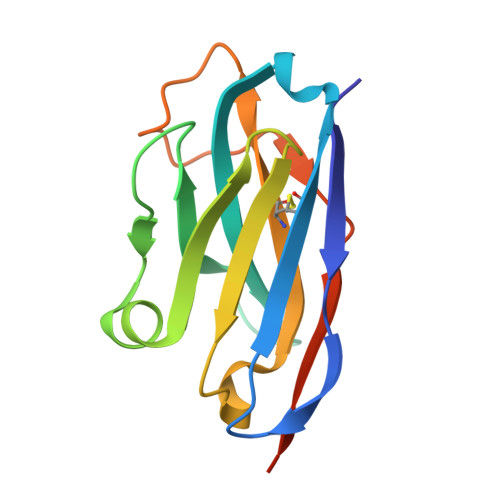
On the humanization of VHHs: Prospective case studies, experimental and computational characterization of structural determinants for functionality.
Fernandez-Quintero, M.L., Guarnera, E., Musil, D., Pekar, L., Sellmann, C., Freire, F., Sousa, R.L., Santos, S.P., Freitas, M.C., Bandeiras, T.M., Silva, M.M.S., Loeffler, J.R., Ward, A.B., Harwardt, J., Zielonka, S., Evers, A.(2024) Protein Sci 33: e5176-e5176
- PubMed: 39422475
- DOI: https://doi.org/10.1002/pro.5176
- Primary Citation of Related Structures:
9FWW, 9FXF - PubMed Abstract:
The humanization of camelid-derived variable domain heavy chain antibodies (VHHs) poses challenges including immunogenicity, stability, and potential reduction of affinity. Critical to this process are complementarity-determining regions (CDRs), Vernier and Hallmark residues, shaping the three-dimensional fold and influencing VHH structure and function. Additionally, the presence of non-canonical disulfide bonds further contributes to conformational stability and antigen binding. In this study, we systematically humanized two camelid-derived VHHs targeting the natural cytotoxicity receptor NKp30. Key structural positions in Vernier and Hallmark regions were exchanged with residues from the most similar human germline sequences. The resulting variants were characterized for binding affinities, yield, and purity. Structural binding modes were elucidated through crystal structure determination and AlphaFold2 predictions, providing insights into differences in binding affinity. Comparative structural and molecular dynamics characterizations of selected variants were performed to rationalize their functional properties and elucidate the role of specific sequence motifs in antigen binding. Furthermore, systematic analyses of next-generation sequencing (NGS) and Protein Data Bank (PDB) data was conducted, shedding light on the functional significance of Hallmark motifs and non-canonical disulfide bonds in VHHs in general. Overall, this study provides valuable insights into the structural determinants governing the functional properties of VHHs, offering a roadmap for their rational design, humanization, and optimization for therapeutic applications.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, USA.
Organizational Affiliation: