Structure of glyoxal oxidase from Fusarium graminearum at 1.28 Angstroms resolution
Fong, J.K., Mathieu, Y., Cordeiro, R.L., Van Petegem, F., Brumer, H.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chromosome 3, complete genome | 661 | Fusarium graminearum PH-1 | Mutation(s): 0 Gene Names: FG11097.1, FGRAMPH1_01T21215 | 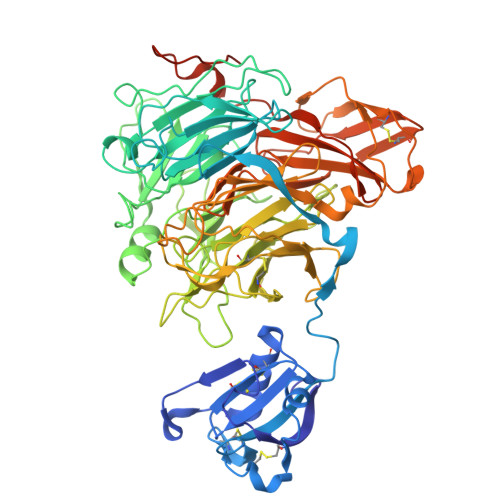 | |
UniProt | |||||
Find proteins for A0A0E0SJJ3 (Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1)) Explore A0A0E0SJJ3 Go to UniProtKB: A0A0E0SJJ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0E0SJJ3 | ||||
Glycosylation | |||||
| Glycosylation Sites: 4 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C [auth B] | 2 |  | N-Glycosylation | |
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | E [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| SO4 Query on SO4 | G [auth A], H [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CU Query on CU | F [auth A] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| NA Query on NA | I [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86 | α = 90 |
| b = 129.98 | β = 90 |
| c = 134.05 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | RGPIN-2018-03892 |
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | RGPIN-2024-04318 |
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | CREATE 509257-18 |