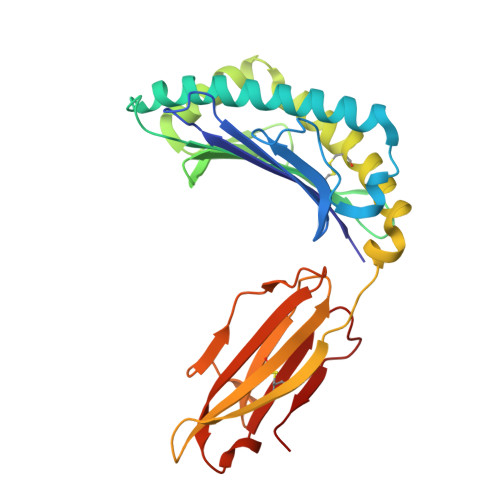
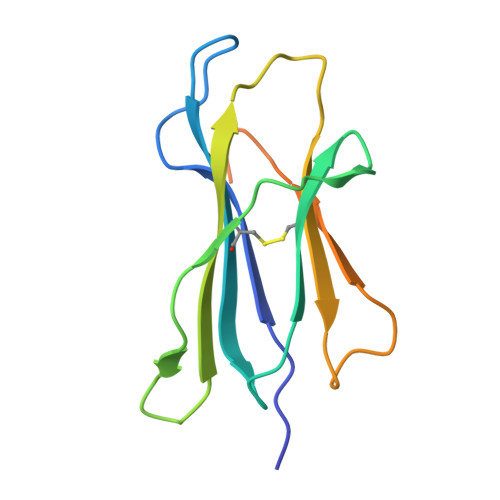
Structure guided analysis of KRAS G12 mutants in HLA-A*11:01 reveals a length encoded immunogenic advantage in G12D.
Zhu, J., Chen, Z., Xu, X., Wang, Y., Liu, P., Wen, M., Wang, Q., He, Y., Jin, H., Xue, H., Wang, S., Xu, K., Zhao, L.(2025) Commun Biol 9: 26-26
- PubMed: 41339521
- DOI: https://doi.org/10.1038/s42003-025-09285-0
- Primary Citation of Related Structures:
8K4T, 8K4V, 8K50, 9UV8, 9WVE, 9WVF - PubMed Abstract:
KRAS G12 mutations are frequent oncogenic drivers, yet their differential immunogenicity complicates T cell-based therapies. Here, we integrate structural, biophysical, and functional analyses to examine how KRAS G12 variants remodel peptide-MHC-I (pMHC) architecture and T cell receptor (TCR) recognition. Using HLA-A*11:01, we show that single residue substitutions at position 12 induce distinct conformational changes in the MHC groove, with G12D uniquely destabilizing the complex through a buried aspartate side chain. Notably, G12D peptides adopt two registers, a 9-mer and a 10-mer, that diverge sharply in structure and immunogenicity. The 10-mer forms a compact, stable pMHC with a TCR-accessible surface, while the 9-mer adopts a bent conformation incompatible with recognition. Molecular dynamics and NMR titration confirm the superior stability and binding affinity of the 10-mer. These results highlight how peptide length and conformation critically shape immune visibility, offering mechanistic insight for optimizing TCR-T therapies against elusive neoantigens like KRAS G12D.
- Department of Anesthesiology, Putuo People's Hospital, School of Medicine, Tongji University, Shanghai, China.
Organizational Affiliation: