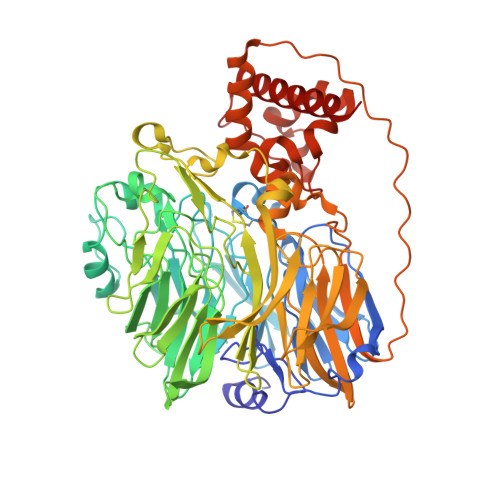
Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Chen, Z.W., Matsushita, K., Yamashita, T., Fujii, T.A., Toyama, H., Adachi, O., Bellamy, H.D., Mathews, F.S.(2002) Structure 10: 837-849
- PubMed: 12057198
- DOI: https://doi.org/10.1016/s0969-2126(02)00774-8
- Primary Citation of Related Structures:
1KV9 - PubMed Abstract:
The type II quinohemoprotein alcohol dehydrogenase of Pseudomonas putida is a periplasmic enzyme that oxidizes substrate alcohols to the aldehyde and transfers electrons first to pyrroloquinoline quinone (PQQ) and then to an internal heme group. The 1.9 A resolution crystal structure reveals that the enzyme contains a large N-terminal eight-stranded beta propeller domain (approximately 60 kDa) similar to methanol dehydrogenase and a small C-terminal c-type cytochrome domain (approximately 10 kDa) similar to the cytochrome subunit of p-cresol methylhydoxylase. The PQQ is bound near the axis of the propeller domain about 14 A from the heme. A molecule of acetone, the product of the oxidation of isopropanol present during crystallization, appears to be bound in the active site cavity.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: