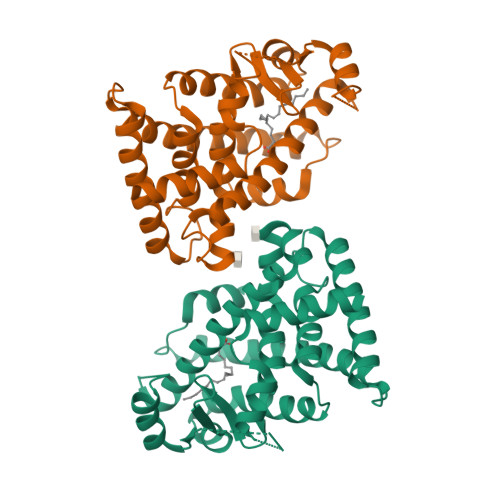
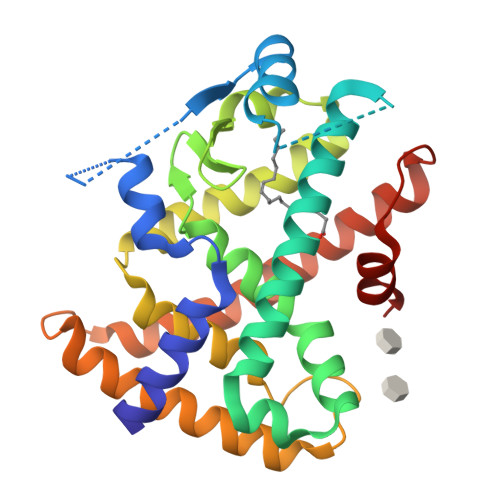
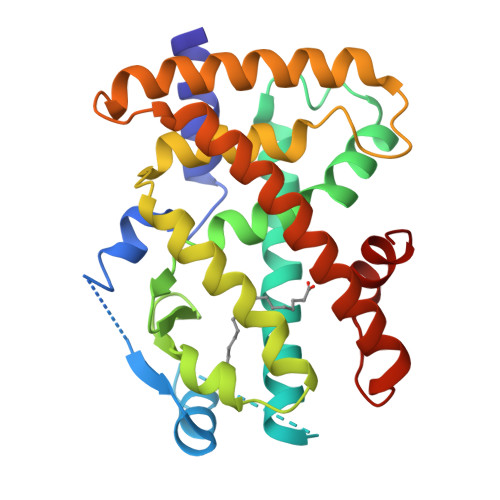
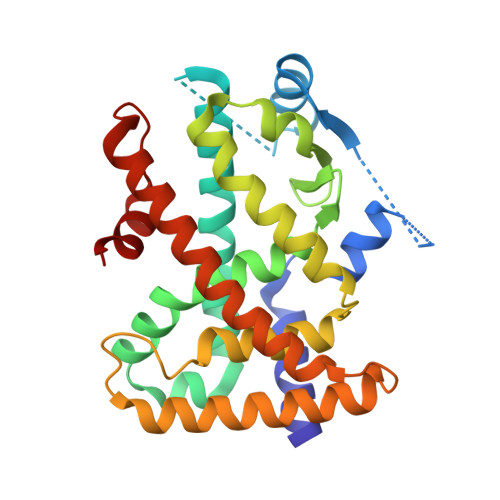
Reevaluation of the PPAR-beta/delta Ligand Binding Domain Model Reveals Why It Exhibits the Activated Form
Fyffe, S.A., Alphey, M.S., Buetow, L., Smith, T.K., Ferguson, M.A.J., Sorensen, M.D., Bjorkling, F., Hunter, W.N.(2006) Mol Cell 21: 1-2
- PubMed: 16387648
- DOI: https://doi.org/10.1016/j.molcel.2005.12.001
- Primary Citation of Related Structures:
2BAW
Organizational Affiliation:
Division of Biological Chemistry and Molecular Microbiology, School of Life Sciences, University of Dundee, Dundee DD1 5EH, United Kingdom.