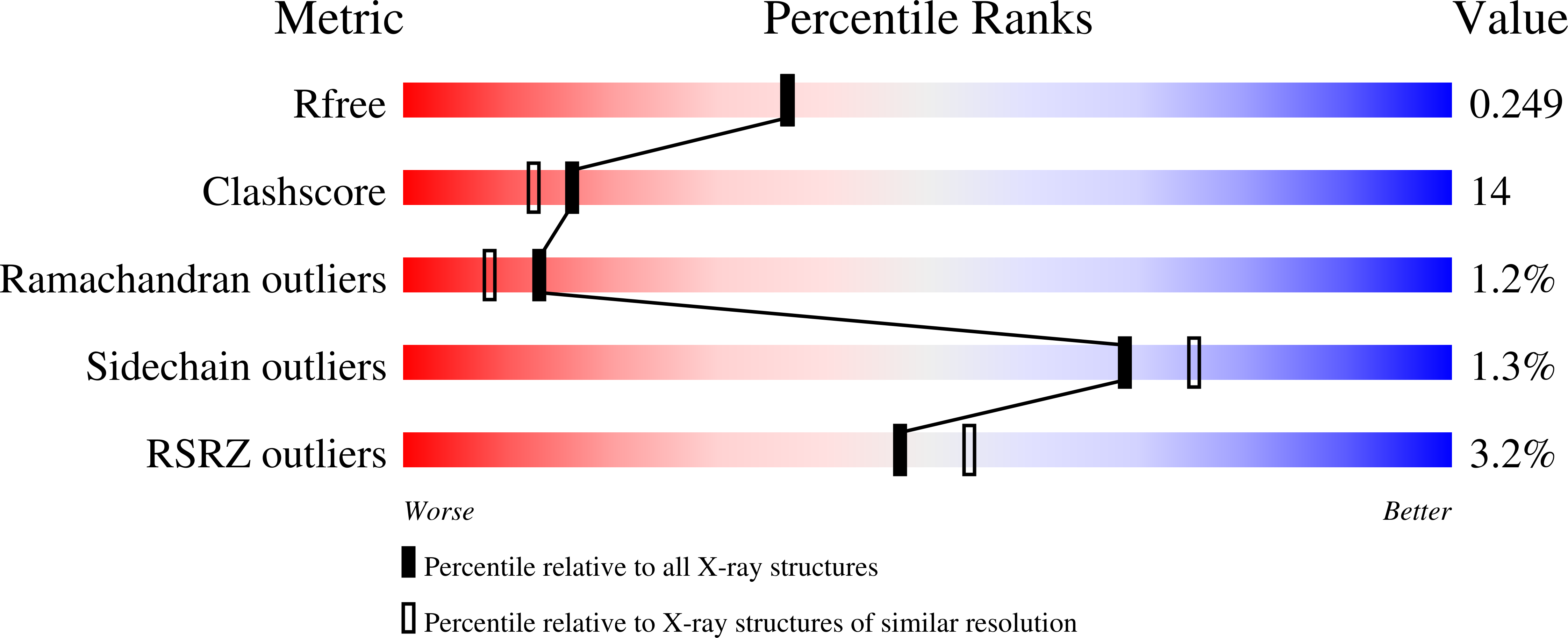
Structural characterization of CA1462, the Candida albicans thiamine pyrophosphokinase.
Santini, S., Monchois, V., Mouz, N., Sigoillot, C., Rousselle, T., Claverie, J.M., Abergel, C.(2008) BMC Struct Biol 8: 33-33
- PubMed: 18652651
- DOI: https://doi.org/10.1186/1472-6807-8-33
- Primary Citation of Related Structures:
2G9Z, 2HH9 - PubMed Abstract:
In search of new antifungal targets of potential interest for pharmaceutical companies, we initiated a comparative genomics study to identify the most promising protein-coding genes in fungal genomes. One criterion was the protein sequence conservation between reference pathogenic genomes. A second criterion was that the corresponding gene in Saccharomyces cerevisiae should be essential. Since thiamine pyrophosphate is an essential product involved in a variety of metabolic pathways, proteins responsible for its production satisfied these two criteria. We report the enzymatic characterization and the crystallographic structure of the Candida albicans Thiamine pyrophosphokinase. The protein was co-crystallized with thiamine or thiamine-PNP. The presence of an inorganic phosphate in the crystallographic structure opposite the known AMP binding site relative to the thiamine moiety suggests that a second AMP molecule could be accommodated in the C. albicans structure. Together with the crystallographic structures of the enzyme/substrate complexes this suggests the existence of a secondary, less specific, nucleotide binding site in the Candida albicans thiamine pyrophosphokinase which could transiently serve during the release or the binding of ATP. The structures also highlight a conserved Glutamine residue (Q138) which could interact with the ATP alpha-phosphate and act as gatekeeper. Finally, the TPK/Thiamine-PNP complex is consistent with a one step mechanism of pyrophosphorylation.
Organizational Affiliation:
Information Genomique et Structurale, UPR2589, Parc Scientifique de Luminy, 13288, Marseille cedex 09, France. santini.s@fsagx.ac.be