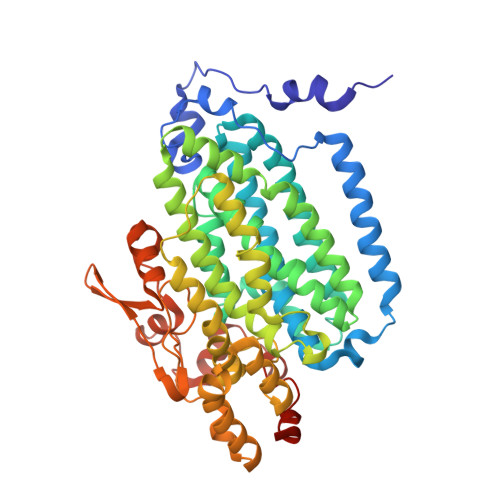
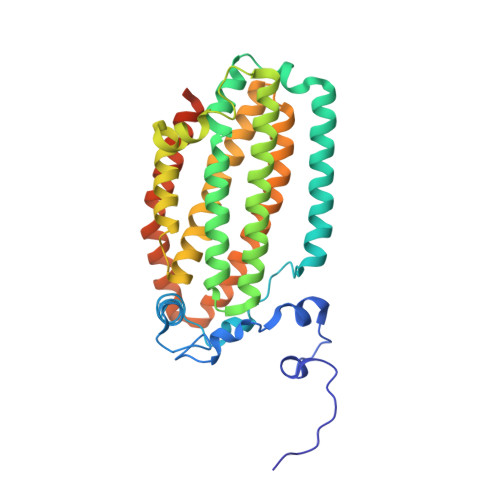
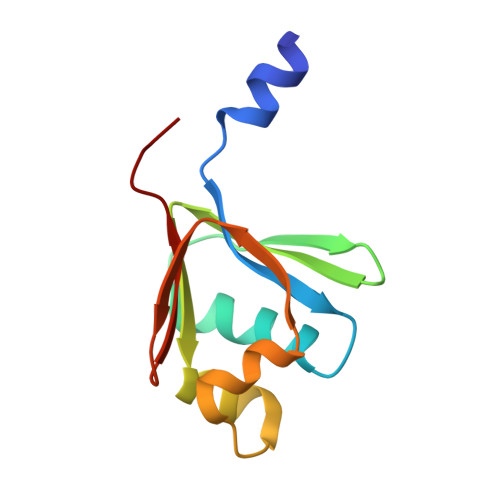
Structural consequences of effector protein complex formation in a diiron hydroxylase.
Bailey, L.J., McCoy, J.G., Phillips Jr., G.N., Fox, B.G.(2008) Proc Natl Acad Sci U S A 105: 19194-19198
- PubMed: 19033467
- DOI: https://doi.org/10.1073/pnas.0807948105
- Primary Citation of Related Structures:
3DHG, 3DHH, 3DHI - PubMed Abstract:
Carboxylate-bridged diiron hydroxylases are multicomponent enzyme complexes responsible for the catabolism of a wide range of hydrocarbons and as such have drawn attention for their mechanism of action and potential uses in bioremediation and enzymatic synthesis. These enzyme complexes use a small molecular weight effector protein to modulate the function of the hydroxylase. However, the origin of these functional changes is poorly understood. Here, we report the structures of the biologically relevant effector protein-hydroxylase complex of toluene 4-monooxygenase in 2 redox states. The structures reveal a number of coordinated changes that occur up to 25 A from the active site and poise the diiron center for catalysis. The results provide a structural basis for the changes observed in a number of the measurable properties associated with effector protein binding. This description provides insight into the functional role of effector protein binding in all carboxylate-bridged diiron hydroxylases.
- Department of Biochemistry, University of Wisconsin, 433 Babcock Drive, Madison, WI 53706-1544, USA.
Organizational Affiliation: