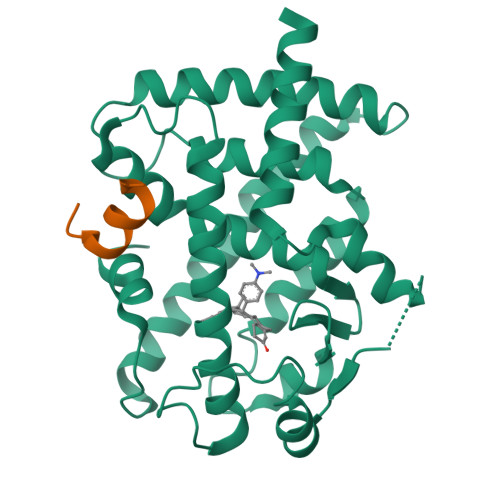
Revealing a steroid receptor ligand as a unique PPAR gamma agonist.
Lin, S., Han, Y., Shi, Y., Rong, H., Zheng, S., Jin, S., Lin, S.Y., Lin, S.C., Li, Y.(2012) Cell Res 22: 746-756
- PubMed: 21986665
- DOI: https://doi.org/10.1038/cr.2011.162
- Primary Citation of Related Structures:
3QT0 - PubMed Abstract:
Peroxisome proliferator-activated receptor gamma (PPARγ) regulates metabolic homeostasis and is a molecular target for anti-diabetic drugs. We report here the identification of a steroid receptor ligand, RU-486, as an unexpected PPARγ agonist, thereby uncovering a novel signaling route for this steroid drug. Similar to rosiglitazone, RU-486 modulates the expression of key PPARγ target genes and promotes adipocyte differentiation, but with a lower adipogenic activity. Structural and functional studies of receptor-ligand interactions reveal the molecular basis for a unique binding mode for RU-486 in the PPARγ ligand-binding pocket with distinctive properties and epitopes, providing the molecular mechanisms for the discrimination of RU-486 from thiazolidinediones (TZDs) drugs. Our findings together indicate that steroid compounds may represent an alternative approach for designing non-TZD PPARγ ligands in the treatment of insulin resistance.
Organizational Affiliation:
State Key Laboratory for Cellular Stress Biology, School of Life Sciences, Xiamen University, Fujian, China.