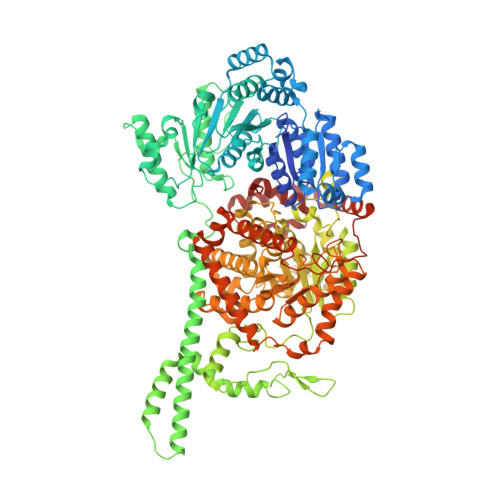
Visualization of a radical B12 enzyme with its G-protein chaperone.
Jost, M., Cracan, V., Hubbard, P.A., Banerjee, R., Drennan, C.L.(2015) Proc Natl Acad Sci U S A 112: 2419-2424
- PubMed: 25675500
- DOI: https://doi.org/10.1073/pnas.1419582112
- Primary Citation of Related Structures:
4XC6, 4XC7, 4XC8 - PubMed Abstract:
G-protein metallochaperones ensure fidelity during cofactor assembly for a variety of metalloproteins, including adenosylcobalamin (AdoCbl)-dependent methylmalonyl-CoA mutase and hydrogenase, and thus have both medical and biofuel development applications. Here, we present crystal structures of IcmF, a natural fusion protein of AdoCbl-dependent isobutyryl-CoA mutase and its corresponding G-protein chaperone, which reveal the molecular architecture of a G-protein metallochaperone in complex with its target protein. These structures show that conserved G-protein elements become ordered upon target protein association, creating the molecular pathways that both sense and report on the cofactor loading state. Structures determined of both apo- and holo-forms of IcmF depict both open and closed enzyme states, in which the cofactor-binding domain is alternatively positioned for cofactor loading and for catalysis. Notably, the G protein moves as a unit with the cofactor-binding domain, providing a visualization of how a chaperone assists in the sequestering of a precious cofactor inside an enzyme active site.
- Department of Chemistry and.
Organizational Affiliation: