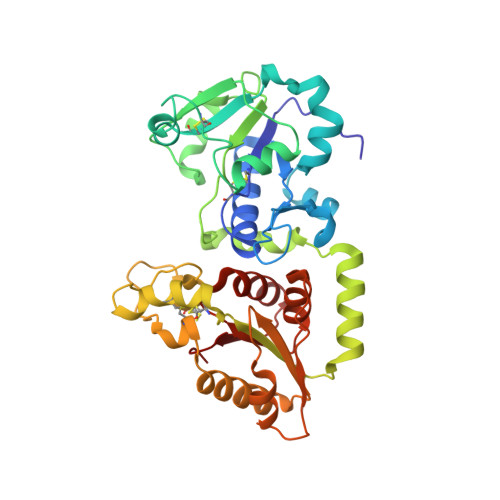
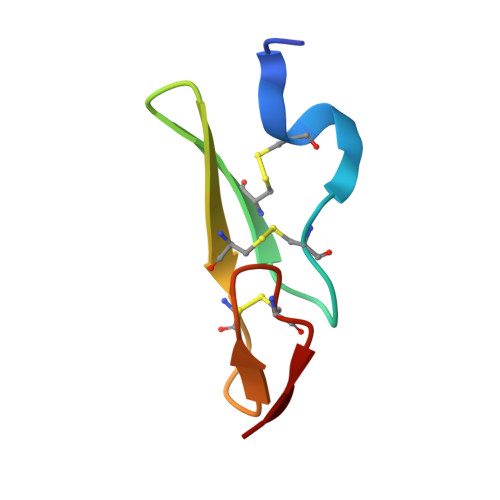
Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Li, Z., Han, K., Pak, J.E., Satkunarajah, M., Zhou, D., Rini, J.M.(2017) Nat Chem Biol 13: 757-763
- PubMed: 28530709
- DOI: https://doi.org/10.1038/nchembio.2381
- Primary Citation of Related Structures:
5KXH, 5KXQ, 5KY0, 5KY2, 5KY3, 5KY4, 5KY5, 5KY7, 5KY8, 5KY9 - PubMed Abstract:
Protein O-fucosyltransferase 1 (POFUT1) fucosylates the epidermal growth factor (EGF)-like domains found in cell-surface and secreted glycoproteins including Notch and its ligands. Although Notch fucosylation is critical for development, and POFUT1 deficiency leads to human disease, how this enzyme binds and catalyzes the fucosylation of its diverse EGF-like domain substrates has not been determined. Reported here is the X-ray crystal structure of mouse POFUT1 in complex with several EGF-like domains, including EGF12 and EGF26 of Notch. Overall shape complementarity, interactions with invariant atoms of the fucosylation motif and flexible segments on POFUT1 all define its EGF-like-domain binding properties. Using large-scale structural and sequence analysis, we also show that POFUT1 binds EGF-like domains of the hEGF type and that the highly correlated presence of POFUT1 and fucosylatable hEGFs has accompanied animal evolution.
- Department of Biochemistry, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: