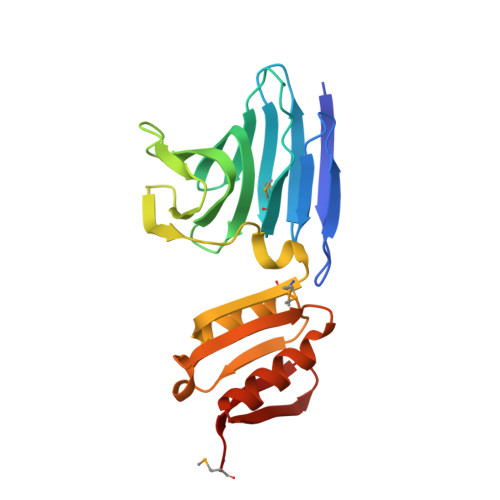
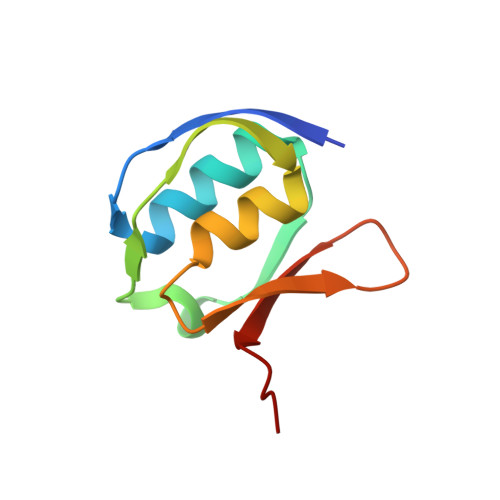
Structural basis of cell-surface signaling by a conserved sigma regulator in Gram-negative bacteria.
Jensen, J.L., Jernberg, B.D., Sinha, S.C., Colbert, C.L.(2020) J Biol Chem 295: 5795-5806
- PubMed: 32107313
- DOI: https://doi.org/10.1074/jbc.RA119.010697
- Primary Citation of Related Structures:
6OVK, 6OVM - PubMed Abstract:
Cell-surface signaling (CSS) in Gram-negative bacteria involves highly conserved regulatory pathways that optimize gene expression by transducing extracellular environmental signals to the cytoplasm via inner-membrane sigma regulators. The molecular details of ferric siderophore-mediated activation of the iron import machinery through a sigma regulator are unclear. Here, we present the 1.56 Å resolution structure of the periplasmic complex of the C-terminal CSS domain (CCSSD) of PupR, the sigma regulator in the Pseudomonas capeferrum pseudobactin BN7/8 transport system, and the N-terminal signaling domain (NTSD) of PupB, an outer-membrane TonB-dependent transducer. The structure revealed that the CCSSD consists of two subdomains: a juxta-membrane subdomain, which has a novel all-β-fold, followed by a secretin/TonB, short N-terminal subdomain at the C terminus of the CCSSD, a previously unobserved topological arrangement of this domain. Using affinity pulldown assays, isothermal titration calorimetry, and thermal denaturation CD spectroscopy, we show that both subdomains are required for binding the NTSD with micromolar affinity and that NTSD binding improves CCSSD stability. Our findings prompt us to present a revised model of CSS wherein the CCSSD:NTSD complex forms prior to ferric-siderophore binding. Upon siderophore binding, conformational changes in the CCSSD enable regulated intramembrane proteolysis of the sigma regulator, ultimately resulting in transcriptional regulation.
Organizational Affiliation:
Department of Chemistry and Biochemistry, North Dakota State University, Fargo, North Dakota 58108.