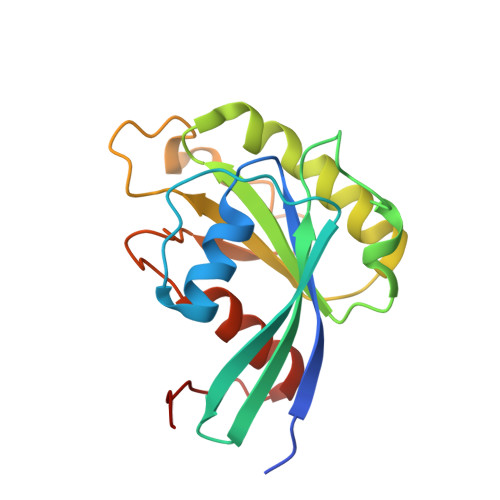
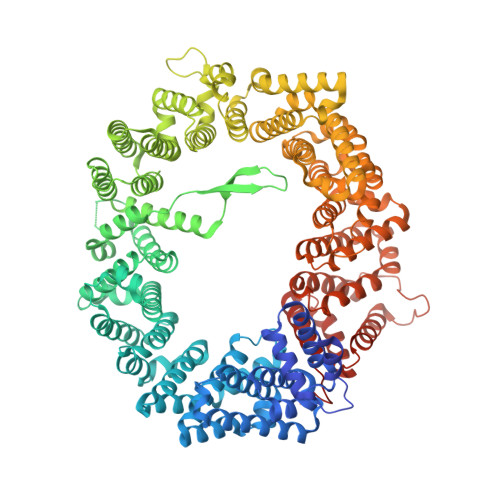
Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
Shaikhqasem, A., Dickmanns, A., Neumann, P., Ficner, R.(2020) J Med Chem 63: 7545-7558
- PubMed: 32585100
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00143
- Primary Citation of Related Structures:
6TVO - PubMed Abstract:
The receptor CRM1 is responsible for the nuclear export of many tumor-suppressor proteins and viral ribonucleoproteins. This renders CRM1 an interesting target for therapeutic intervention in diverse cancer types and viral diseases. Structural studies of Saccharomyces cerevisiae CRM1 ( Sc CRM1) complexes with inhibitors defined the molecular basis for CRM1 inhibition. Nevertheless, no structural information is available for inhibitors bound to human CRM1 ( Hs CRM1). Here, we present the structure of the natural inhibitor Leptomycin B bound to the Hs CRM1-RanGTP complex. Despite high sequence conservation and structural similarity in the NES-binding cleft region, Sc CRM1 exhibits 16-fold lower binding affinity than Hs CRM1 toward PKI-NES and significant differences in affinities toward potential CRM1 inhibitors. In contrast to Hs CRM1, competition assays revealed that a human adapted mutant Sc CRM1-T539C does not bind all inhibitors tested. Taken together, our data indicate the importance of using Hs CRM1 for molecular analysis and development of novel antitumor and antiviral drugs.
- Department of Molecular Structural Biology, Institute of Microbiology and Genetics, GZMB, Georg-August-University Göttingen, 37077 Göttingen, Germany.
Organizational Affiliation: