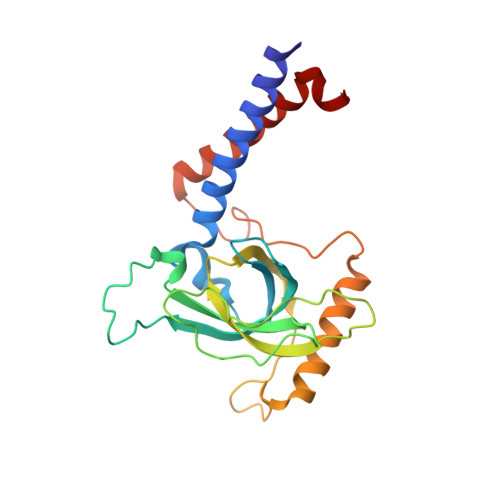
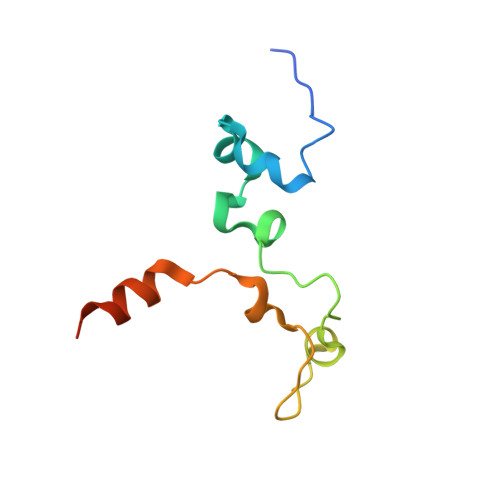
The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Dobrev, N., Ahmed, Y.L., Sivadas, A., Soni, K., Fischer, T., Sinning, I.(2021) Nat Commun 12: 3456-3456
- PubMed: 34103492
- DOI: https://doi.org/10.1038/s41467-021-23565-3
- Primary Citation of Related Structures:
6YFV, 6YGU - PubMed Abstract:
Cryptic unstable transcripts (CUTs) are rapidly degraded by the nuclear exosome in a process requiring the RNA helicase Mtr4 and specific adaptor complexes for RNA substrate recognition. The PAXT and MTREC complexes have recently been identified as homologous exosome adaptors in human and fission yeast, respectively. The eleven-subunit MTREC comprises the zinc-finger protein Red1 and the Mtr4 homologue Mtl1. Here, we use yeast two-hybrid and pull-down assays to derive a detailed interaction map. We show that Red1 bridges MTREC submodules and serves as the central scaffold. In the crystal structure of a minimal Mtl1/Red1 complex an unstructured region adjacent to the Red1 zinc-finger domain binds to both the Mtl1 KOW domain and stalk helices. This interaction extends the canonical interface seen in Mtr4-adaptor complexes. In vivo mutational analysis shows that this interface is essential for cell survival. Our results add to Mtr4 versatility and provide mechanistic insights into the MTREC complex.
Organizational Affiliation:
Heidelberg University Biochemistry Center (BZH), Heidelberg, Germany.