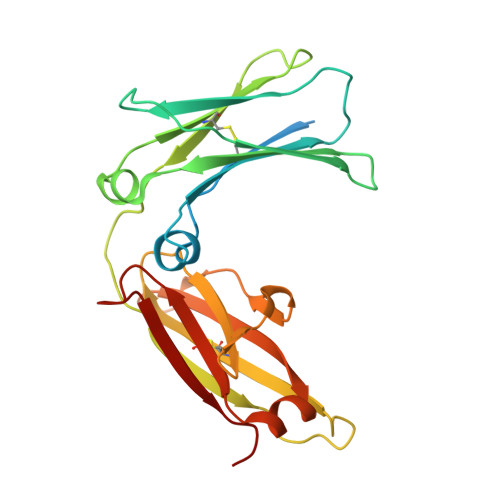
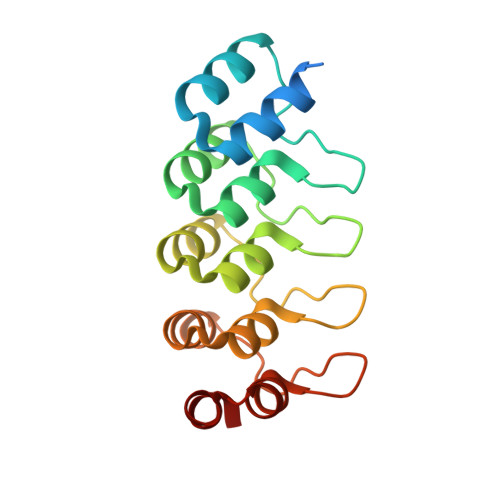
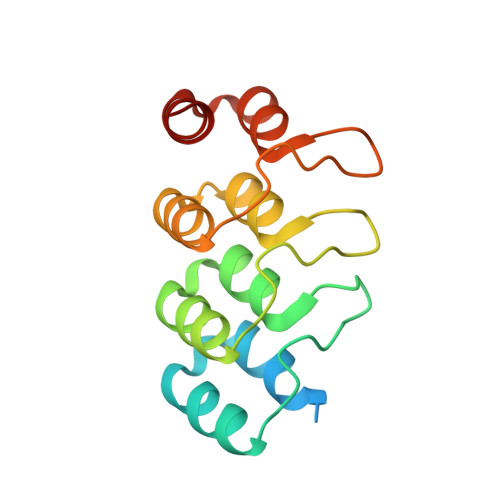
Structure-guided design of ultrapotent disruptive IgE inhibitors to rapidly terminate acute allergic reactions.
Pennington, L.F., Gasser, P., Brigger, D., Guntern, P., Eggel, A., Jardetzky, T.S.(2021) J Allergy Clin Immunol 148: 1049-1060
- PubMed: 33991582
- DOI: https://doi.org/10.1016/j.jaci.2021.03.050
- Primary Citation of Related Structures:
7MXI - PubMed Abstract:
Anaphylaxis represents one of the most severe and fatal forms of allergic reactions. Like most other allergies, it is caused by activation of basophils and mast cells by allergen-mediated cross-linking of IgE bound to its high-affinity receptor, FcεRI, on the cell surface. The systemic release of soluble mediators induces an inflammatory cascade, rapidly causing symptoms with peak severity in minutes to hours after allergen exposure. Primary treatment for anaphylaxis consists of immediate intramuscular administration of adrenaline.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, Calif; Program in Immunology, Stanford University School of Medicine, Stanford, Calif; Sean N. Parker Center for Allergy Research at Stanford University, Stanford, Calif.