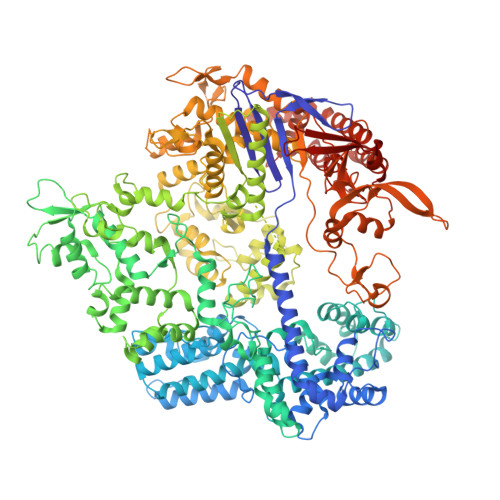
R-loop formation and conformational activation mechanisms of Cas9.
Pacesa, M., Loeff, L., Querques, I., Muckenfuss, L.M., Sawicka, M., Jinek, M.(2022) Nature 609: 191-196
- PubMed: 36002571
- DOI: https://doi.org/10.1038/s41586-022-05114-0
- Primary Citation of Related Structures:
7Z4C, 7Z4D, 7Z4E, 7Z4G, 7Z4H, 7Z4I, 7Z4J, 7Z4K, 7Z4L - PubMed Abstract:
Cas9 is a CRISPR-associated endonuclease capable of RNA-guided, site-specific DNA cleavage 1-3 . The programmable activity of Cas9 has been widely utilized for genome editing applications 4-6 , yet its precise mechanisms of target DNA binding and off-target discrimination remain incompletely understood. Here we report a series of cryo-electron microscopy structures of Streptococcus pyogenes Cas9 capturing the directional process of target DNA hybridization. In the early phase of R-loop formation, the Cas9 REC2 and REC3 domains form a positively charged cleft that accommodates the distal end of the target DNA duplex. Guide-target hybridization past the seed region induces rearrangements of the REC2 and REC3 domains and relocation of the HNH nuclease domain to assume a catalytically incompetent checkpoint conformation. Completion of the guide-target heteroduplex triggers conformational activation of the HNH nuclease domain, enabled by distortion of the guide-target heteroduplex, and complementary REC2 and REC3 domain rearrangements. Together, these results establish a structural framework for target DNA-dependent activation of Cas9 that sheds light on its conformational checkpoint mechanism and may facilitate the development of novel Cas9 variants and guide RNA designs with enhanced specificity and activity.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, Zurich, Switzerland.