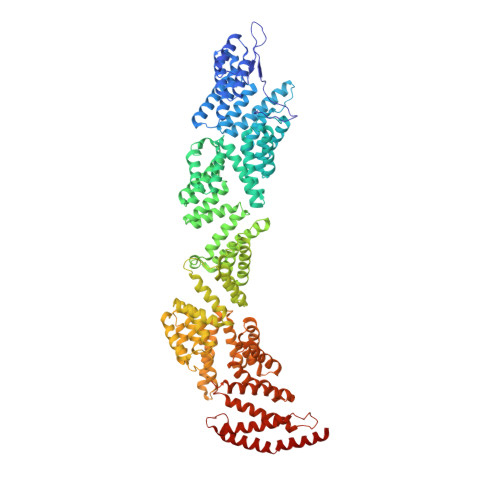
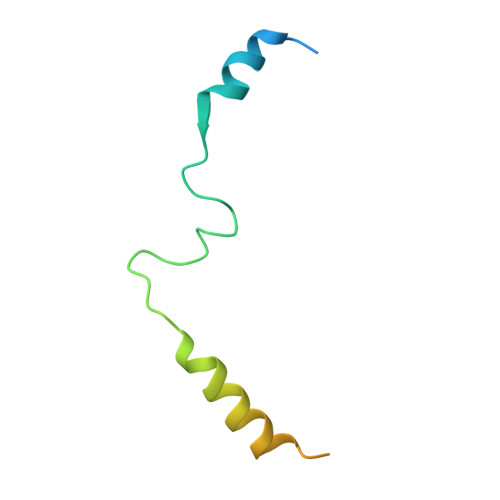
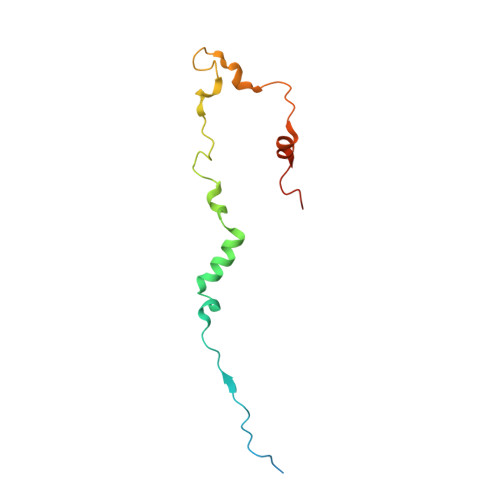
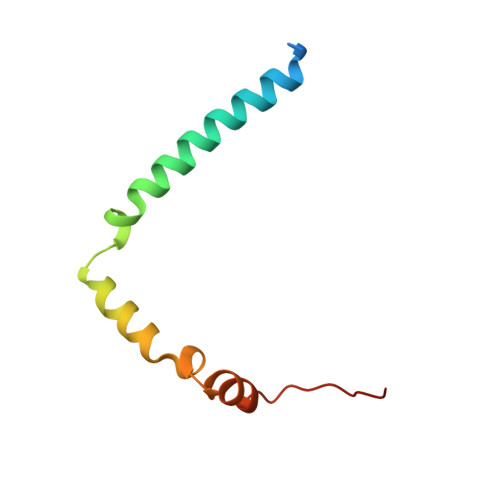
Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Qin, Y., Zhou, Y., Cao, Y., Ren, Y., Deng, P., Jiang, J., Wang, Z.(2023) Int J Mol Sci 24
- PubMed: 37240075
- DOI: https://doi.org/10.3390/ijms24108730
- Primary Citation of Related Structures:
8J8P, 8J8Q - PubMed Abstract:
The multicomponent polymerase associated factor 1 (Paf1) complex (PAF1C) is an important transcription elongation factor that upregulates RNA polymerase II-mediated genome-wide transcription. PAF1C can regulate transcription through direct association with the polymerase or by impacting the chromatin structure epigenetically. In recent years, significant progress has been made in understanding the molecular mechanisms of PAF1C. However, high-resolution structures that can clarify the interaction details among the components of the complex are still needed. In this study, we evaluated the structural core of the yeast PAF1C containing the four components Ctr9, Paf1, Cdc73 and Rtf1 at high resolution. We observed the interaction details among these components. In particular, we identified a new binding surface of Rtf1 on PAF1C and found that the C-terminal sequence of Rtf1 dramatically changed during evolution, which may account for its different binding affinities to PAF1C among species. Our work presents a precise model of PAF1C, which will facilitate our understanding of the molecular mechanism and the in vivo function of the yeast PAF1C.
Organizational Affiliation:
Key Laboratory of Cell Proliferation and Regulation Biology of Ministry of Education, College of Life Sciences, Beijing Normal University, 19 Xinjiekouwai Avenue, Beijing 100875, China.