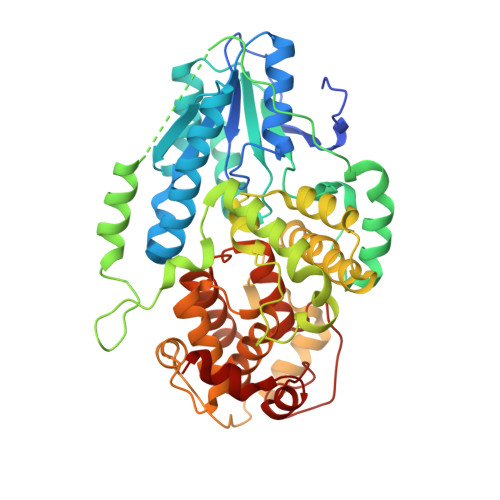
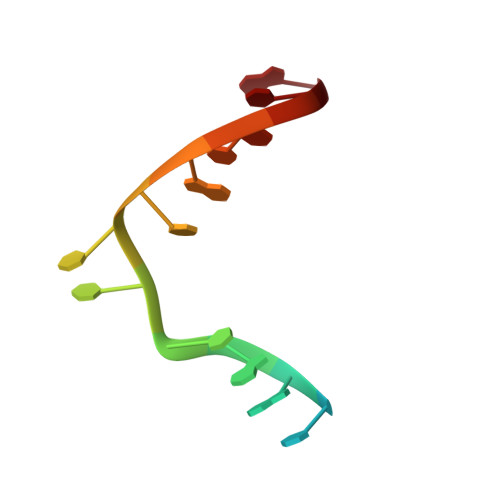
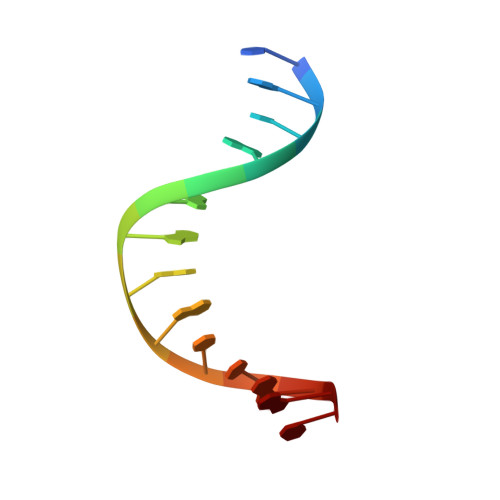
Visualizing the DNA repair process by a photolyase at atomic resolution.
Maestre-Reyna, M., Wang, P.H., Nango, E., Hosokawa, Y., Saft, M., Furrer, A., Yang, C.H., Gusti Ngurah Putu, E.P., Wu, W.J., Emmerich, H.J., Caramello, N., Franz-Badur, S., Yang, C., Engilberge, S., Wranik, M., Glover, H.L., Weinert, T., Wu, H.Y., Lee, C.C., Huang, W.C., Huang, K.F., Chang, Y.K., Liao, J.H., Weng, J.H., Gad, W., Chang, C.W., Pang, A.H., Yang, K.C., Lin, W.T., Chang, Y.C., Gashi, D., Beale, E., Ozerov, D., Nass, K., Knopp, G., Johnson, P.J.M., Cirelli, C., Milne, C., Bacellar, C., Sugahara, M., Owada, S., Joti, Y., Yamashita, A., Tanaka, R., Tanaka, T., Luo, F., Tono, K., Zarzycka, W., Muller, P., Alahmad, M.A., Bezold, F., Fuchs, V., Gnau, P., Kiontke, S., Korf, L., Reithofer, V., Rosner, C.J., Seiler, E.M., Watad, M., Werel, L., Spadaccini, R., Yamamoto, J., Iwata, S., Zhong, D., Standfuss, J., Royant, A., Bessho, Y., Essen, L.O., Tsai, M.D.(2023) Science 382: eadd7795-eadd7795
- PubMed: 38033054
- DOI: https://doi.org/10.1126/science.add7795
- Primary Citation of Related Structures:
7YC7, 7YCM, 7YCP, 7YCR, 7YD6, 7YD7, 7YD8, 7YDZ, 7YE0, 7YEB, 7YEC, 7YEE, 7YEI, 7YEJ, 7YEK, 7YEL, 7YEM, 8KCM - PubMed Abstract:
Photolyases, a ubiquitous class of flavoproteins, use blue light to repair DNA photolesions. In this work, we determined the structural mechanism of the photolyase-catalyzed repair of a cyclobutane pyrimidine dimer (CPD) lesion using time-resolved serial femtosecond crystallography (TR-SFX). We obtained 18 snapshots that show time-dependent changes in four reaction loci. We used these results to create a movie that depicts the repair of CPD lesions in the picosecond-to-nanosecond range, followed by the recovery of the enzymatic moieties involved in catalysis, completing the formation of the fully reduced enzyme-product complex at 500 nanoseconds. Finally, back-flip intermediates of the thymine bases to reanneal the DNA were captured at 25 to 200 microseconds. Our data cover the complete molecular mechanism of a photolyase and, importantly, its chemistry and enzymatic catalysis at work across a wide timescale and at atomic resolution.
- Institute of Biological Chemistry, Academia Sinica, 128 Academia Rd. Sec. 2, Nankang, Taipei 115, Taiwan.
Organizational Affiliation: