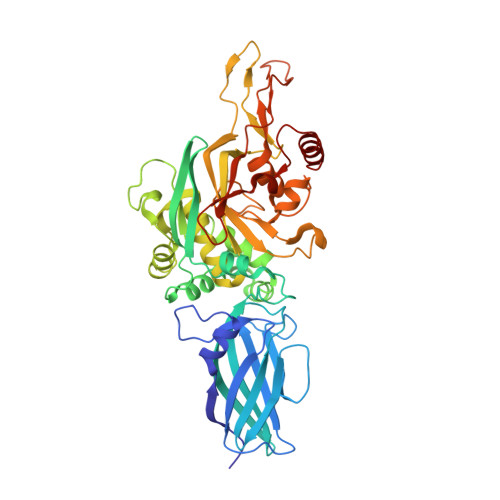
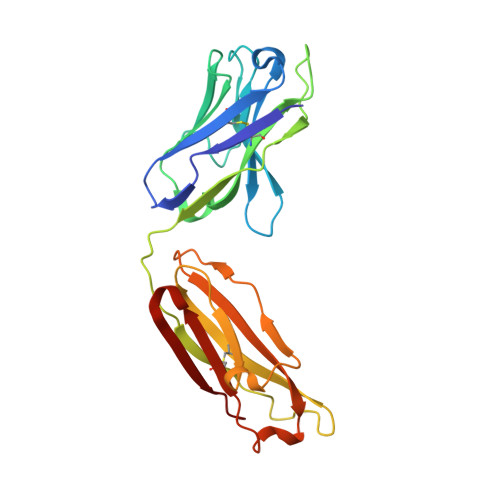
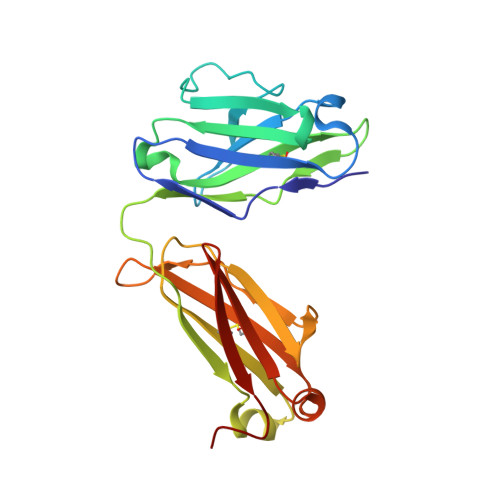
Autoantibody binding and unique enzyme-substrate intermediate conformation of human transglutaminase 3.
Heggelund, J.E., Das, S., Stamnaes, J., Iversen, R., Sollid, L.M.(2023) Nat Commun 14: 6216-6216
- PubMed: 37798283
- DOI: https://doi.org/10.1038/s41467-023-42004-z
- Primary Citation of Related Structures:
8OXV, 8OXW, 8OXX, 8OXY - PubMed Abstract:
Transglutaminase 3 (TG3), the autoantigen of dermatitis herpetiformis (DH), is a calcium dependent enzyme that targets glutamine residues in polypeptides for either transamidation or deamidation modifications. To become catalytically active TG3 requires proteolytic cleavage between the core domain and two C-terminal β-barrels (C1C2). Here, we report four X-ray crystal structures representing inactive and active conformations of human TG3 in complex with a TG3-specific Fab fragment of a DH patient derived antibody. We demonstrate that cleaved TG3, upon binding of a substrate-mimicking inhibitor, undergoes a large conformational change as a β-sheet in the catalytic core domain moves and C1C2 detaches. The unique enzyme-substrate conformation of TG3 without C1C2 is recognized by DH autoantibodies. The findings support a model where B-cell receptors of TG3-specific B cells bind and internalize TG3-gluten enzyme-substrate complexes thereby facilitating gluten-antigen presentation, T-cell help and autoantibody production.
Organizational Affiliation:
KG Jebsen Coeliac Disease Research Centre, Institute of Clinical Medicine, University of Oslo, Oslo, Norway. j.e.heggelund@medisin.uio.no.