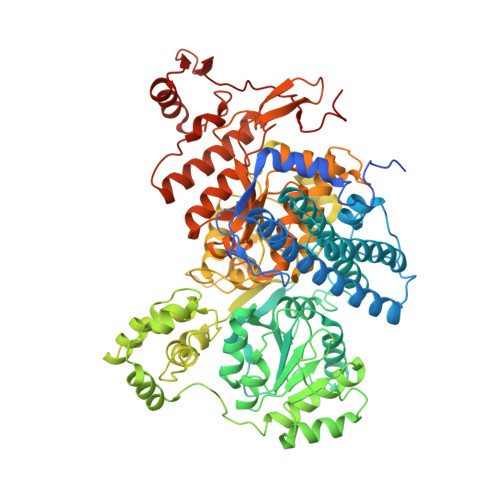
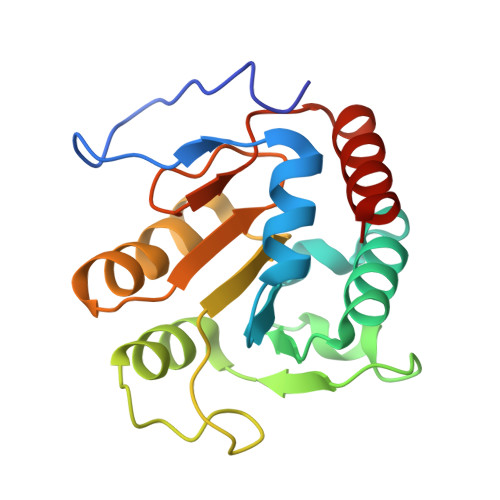
Capturing a methanogenic carbon monoxide dehydrogenase/acetyl-CoA synthase complex via cryogenic electron microscopy.
Biester, A., Grahame, D.A., Drennan, C.L.(2024) Proc Natl Acad Sci U S A 121: e2410995121-e2410995121
- PubMed: 39361653
- DOI: https://doi.org/10.1073/pnas.2410995121
- Primary Citation of Related Structures:
9C0Q, 9C0R, 9C0S, 9C0T - PubMed Abstract:
Approximately two-thirds of the estimated one-billion metric tons of methane produced annually by methanogens is derived from the cleavage of acetate. Acetate is broken down by a Ni-Fe-S-containing A-cluster within the enzyme acetyl-CoA synthase (ACS) to carbon monoxide (CO) and a methyl group (CH 3 + ). The methyl group ultimately forms the greenhouse gas methane, whereas CO is converted to the greenhouse gas carbon dioxide (CO 2 ) by a Ni-Fe-S-containing C-cluster within the enzyme carbon monoxide dehydrogenase (CODH). Although structures have been solved of CODH/ACS from acetogens, which use these enzymes to make acetate from CO 2 , no structure of a CODH/ACS from a methanogen has been reported. In this work, we use cryo-electron microscopy to reveal the structure of a methanogenic CODH and CODH/ACS from Methanosarcina thermophila ( Met CODH/ACS). We find that the N-terminal domain of acetogenic ACS, which is missing in all methanogens, is replaced by a domain of CODH. This CODH domain provides a channel for CO to travel between the two catalytic Ni-Fe-S clusters. It generates the binding surface for ACS and creates a remarkably similar CO alcove above the A-cluster using residues from CODH rather than ACS. Comparison of our Met CODH/ACS structure with our Met CODH structure reveals a molecular mechanism to restrict gas flow from the CO channel when ACS departs, preventing CO escape into the cell. Overall, these long-awaited structures of a methanogenic CODH/ACS reveal striking functional similarities to their acetogenic counterparts despite a substantial difference in domain organization.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139.