Efficient and scalable protein design using a relaxed sequence space
Frank, C.J., Dietz, H.(2024) Science
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
(2024) Science
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DE NOVO PROTEIN P600 | 609 | synthetic construct | Mutation(s): 0 | 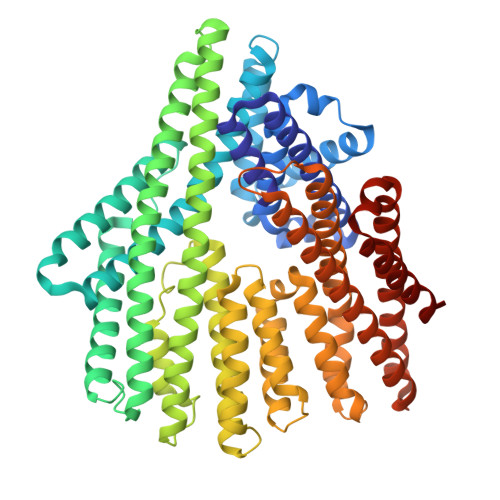 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 163.249 | α = 90 |
| b = 67.825 | β = 103.2 |
| c = 173.693 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Research Council (ERC) | European Union | GA101018465 |
| Germanys Excellence Strategy | Germany | TUM Innovation Network Projekt RISE |