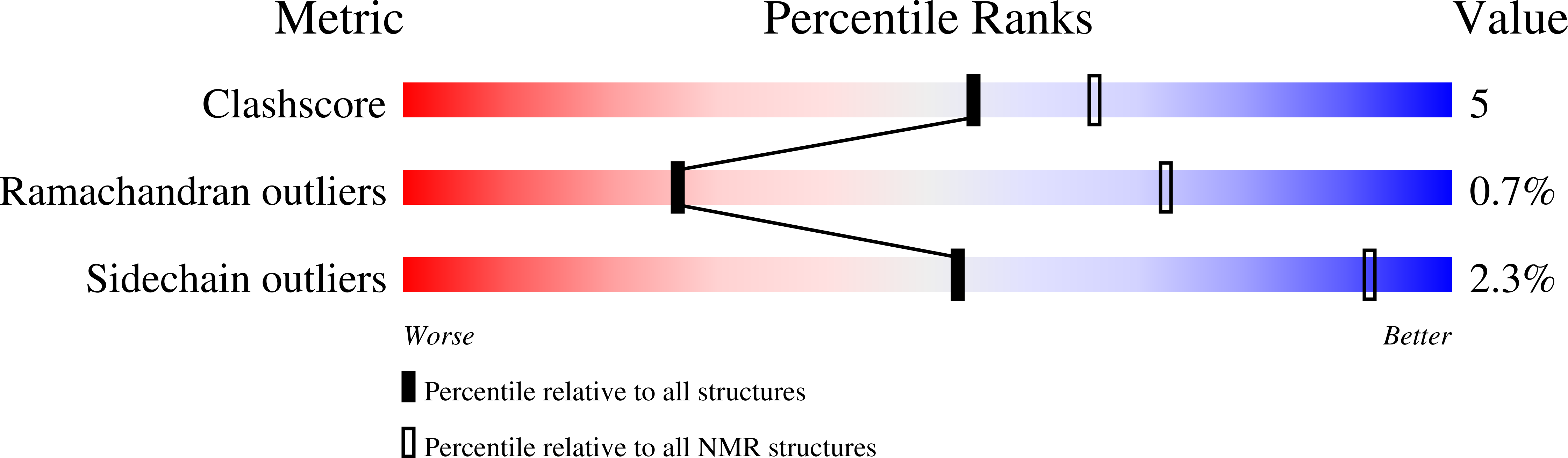Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Belyy, A., Lindemann, F., Roderer, D., Funk, J., Bardiaux, B., Protze, J., Bieling, P., Oschkinat, H., Raunser, S.(2022) Nat Commun 13: 4202-4202
- PubMed: 35858890
- DOI: https://doi.org/10.1038/s41467-022-31836-w
- Primary Citation of Related Structures:
7Z7H, 7Z7I, 7ZBQ - PubMed Abstract:
Tc toxins deliver toxic enzymes into host cells by a unique injection mechanism. One of these enzymes is the actin ADP-ribosyltransferase TccC3, whose activity leads to the clustering of the cellular cytoskeleton and ultimately cell death. Here, we show in atomic detail how TccC3 modifies actin. We find that the ADP-ribosyltransferase does not bind to G-actin but interacts with two consecutive actin subunits of F-actin. The binding of TccC3 to F-actin occurs via an induced-fit mechanism that facilitates access of NAD + to the nucleotide binding pocket. The following nucleophilic substitution reaction results in the transfer of ADP-ribose to threonine-148 of F-actin. We demonstrate that this site-specific modification of F-actin prevents its interaction with depolymerization factors, such as cofilin, which impairs actin network turnover and leads to steady actin polymerization. Our findings reveal in atomic detail a mechanism of action of a bacterial toxin through specific targeting and modification of F-actin.
Organizational Affiliation:
Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Otto-Hahn-Str. 11, 44227, Dortmund, Germany.