New Features
Calculate Pairwise Structure Alignments
03/09
Visualization of structure alignments is a valuable tool for the detection of functionally-related positions in proteins, analysis of conformational changes in ligand binding, exploration of structural variation in protein within an evolutionary family, and identification of common structural domains.
A new pairwise structure alignment interface can be used to align a pair of protein chains using the FATCAT, CE, TM-align and Smith-Waterman 3D algorithms. Chains can be easily selected from structures in the PDB structures or from uploaded coordinate files
Superposed structures will be aligned and launched in the interactive Mol* viewer. Detailed documentation is available.
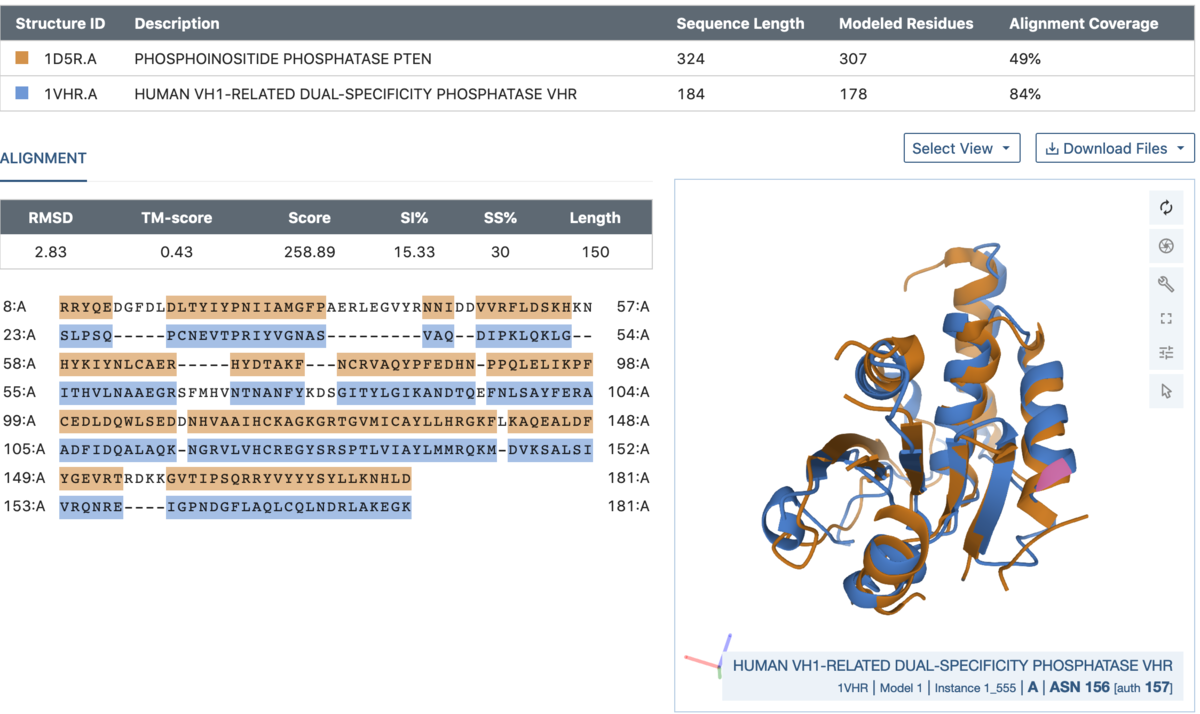 Comparison between chains A of a PTEN tumor suppressor (1D5R.A, blue) and a dual specificity phosphatase (1VHR.A, orange).
Comparison between chains A of a PTEN tumor suppressor (1D5R.A, blue) and a dual specificity phosphatase (1VHR.A, orange).Access the Pairwise Structure Alignment tool from the Analyze menu at the top of the page or directly from RCSB.org/alignment.












