News
Impact of the Protein Data Bank Across Scientific Disciplines
06/30 
 Natalie Verdiguel and Zukang Feng at the 2018 RISE at Rutgers Symposium
Natalie Verdiguel and Zukang Feng at the 2018 RISE at Rutgers SymposiumThis study is the result of a research performed by Natalie Verdiguel (University of Central Florida) during her Research Intensive Summer Experience (RISE) at Rutgers. Working with RCSB PDB developer Zukang Feng, Natalie designed this analysis of the set of data citation information.
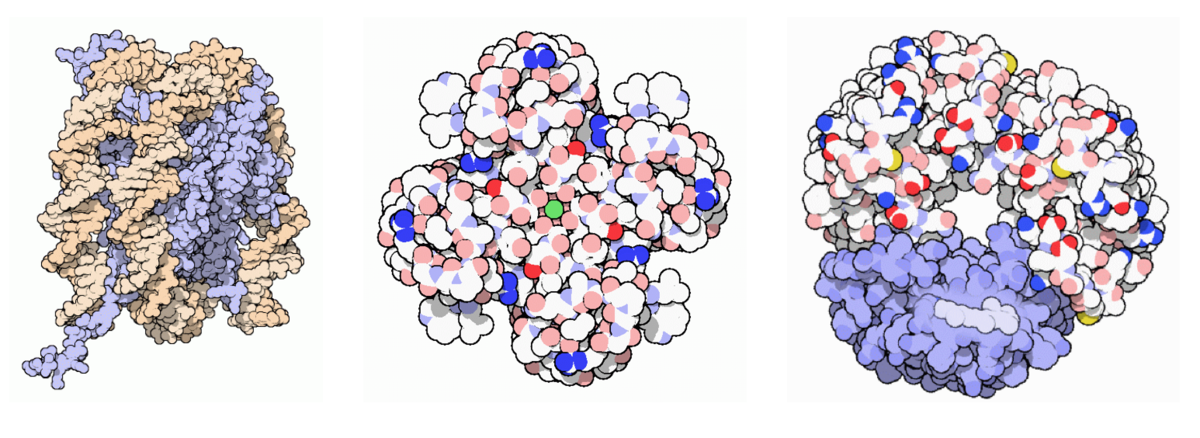
They found that nucleosome structure 1aoi was the most-cited structure in the archive overall, followed by potassium channel (1bl8), bacteriorhodopsin (1brd), rhodopsin (1f88), major histocompatibility class I (1hla), MDM2/imidazoline inhibitor (1rv1), thrombin (2v3o/2v3h), serum albumin (1uor), and ATP Synthase F1 (1bmf).
For more, see Impact of the Protein Data Bank Across Scientific Disciplines Zukang Feng, Natalie Verdiguel, Luigi Di Costanzo, David S. Goodsell, John D. Westbrook, Stephen Burley, and Christine Zardecki (2020) Data Science Journal 19: 25 doi: 10.5334/dsj-2020-025.
The 2020 RISE at Rutgers scholars are working remotely with Rutgers faculty this summer. Students working with the RCSB PDB are focusing on SARS-CoV-2 structures in the PDB.