RCSB PDB Help
Search and Browse > Advanced Search
3D Similarity Search
Introduction
The functions of biological molecules are determined by their 3D shape, which means that molecules with similar 3D structures often share similar functions.
The Protein Data Bank (PDB) continues to grow each year, with an increasing number of experimental and integrative structures of varying size and complexity. Many of these structures represent assemblies, which may consist of multiple proteins or multiple copies of the same protein. The coordinates for these assemblies may include:
- Deposited coordinates or specific subsets of the model
- Coordinates derived from symmetry operations applied to the deposited model
When comparing the shapes of complexes, it is important to consider the full reconstructed assembly, since a single PDB entry may include multiple biologically relevant assemblies.
In addition, RCSB.org provides access to over a million Computed Structure Models (CSMs). Unlike experimental structures, the coordinates of CSMs do not include symmetry-related information, so the model and assembly coordinates are identical.
Finding and classifying 3D structures is essential for understanding functional and evolutionary relationships. While sequence-based searches can identify conserved domains in proteins, many biological examples show that proteins can have similar shapes and functions despite sequence variations. Additionally, a single protein may adopt multiple conformations, such as open and closed forms of an enzyme, which cannot be detected through sequence-based searches alone. These cases require structure similarity search methods.
Many proteins are also stabilized or function as part of assemblies, interacting with one or more copies of themselves or with other proteins. Structure similarity searches allow you to identify similar assemblies, enabling exploration of both the shape of individual proteins and their interactions within complexes.
How 3D Similarity Search Works?
The 3D Similarity search option allows you to query the PDB archive using the three-dimensional (3D) shape of a protein structure. This method, developed by RCSB PDB (Guzenko et al., 2020), represents proteins as volumes of space filled by atoms—focusing on density distribution rather than just atomic coordinates and chain connectivities.
Protein volumes are analyzed using 3D Zernike polynomials and described as vectors of Zernike moments, which provide compact descriptors of the shape. These descriptors are invariant to rotation and translation (Novotni and Klein, 2004), allowing the method to capture the global 3D structure efficiently.
The search uses BioZernike descriptors to assess overall shape similarity and performs rapidly for both individual protein chains and assemblies, enabling fast and accurate identification of structurally similar proteins.
Documentation
You can access the 3D Similarity search by opening Advanced Search and clicking on (+) 3D Similarity from the list of available search tools, or go directly to the search using this link: 3D Similarity Search.
How to Provide a Query
The 3D Similarity search three ways to provide a query structure:
Select an existing PDB or CSM structure
Choose a structure directly from the PDB archive or available Computed Structure Models (CSMs). The selected structure will be loaded automatically for use as the query.
Upload a local coordinates file
Upload a file in a supported format (e.g., PDB, mmCIF, or binaryCIF) from your computer. The uploaded coordinates will be loaded automatically for use as the query.
Files with the extensions “.cif”, “.bcif”, “.pdb”, and “.ent”, as well as their gzipped (“.gz”) versions, are supported. After you select a file, it is automatically uploaded to RCSB PDB servers. Your file will be assigned a unique, randomly generated URL. This URL cannot be guessed by other users; however, anyone who does have the link will be able to access the file.
Uploaded files remain available for 90 days, allowing you to bookmark your search or share it with collaborators during that time. If you need a persistent reference—for example, in a publication, blog post, or any long-lived resource—you should upload your structure to an external file-sharing service (such as Dropbox or Google Drive) and use the URL option instead. This same approach is required for queries saved in MyPDB.
The maximum supported file size is 10 MB. Larger files must be hosted externally and referenced through a URL.
Use URL to reference coordinates file
Provide a direct link to a structure file hosted online. You will need to click Load for the system to retrieve and load the structure for use as the query.
This option can be used to search for structures similar to a 3D model hosted outside of RCSB.org, such as predictions from AlphaFold, RoseTTAFold, or ESMFold, as well as structures available in other public data resources. By providing a direct URL to the coordinates file, the 3D Similarity search will retrieve and load the model automatically, allowing you to use it as the query without needing to download and re-upload the file manually.
Modifying the Query
Once a structure is loaded using any of the three modes, you can refine the query in several ways:
- Use a specific chain or assembly as the query:
- Select a chain by its ID or an assembly by its ID.
- This allows focusing the search on a specific part of the structure rather than the entire molecule.
- Interactive 3D selection:
- Integration with the 3D viewer enables you to visually select the query chain or assembly directly in the structure.
- This provides a more intuitive way to define the search query for complex structures.
Search Options
Precision Mode
For any structure similarity search it is possible to choose between two modes of matching using the drop down menu:
- Strict: Returns only the most reliable matches, but may miss more distant similarities.
- Relaxed: Returns a broader set of similar structures, but may include more false positives.
Search Targets
Controls whether your query structure is compared against individual chains or full assemblies
- Assemblies: Use this to match your query to complete assemblies (this is relevant if you are interested in the overall shape of a complex).
- Chains: Use this to match your query to individual chains of protein structures.
Reasonable defaults are applied automatically. For example, if the query is defined using an assembly, the search will look for assemblies; if a chain is selected, the search will target individual chains.
However, it may be helpful to adjust these options if your query returns no results or does not produce the expected matches.
Query By Example
All 3D structures available from the RCSB.org (experimental structures and CSMs) have a dedicated Structure Summary page that displays information about the entities and assemblies of that entry. To search for structures similar to any one polymer entity in the structure click on the 3D Structure link above the details listed for the macromolecule.
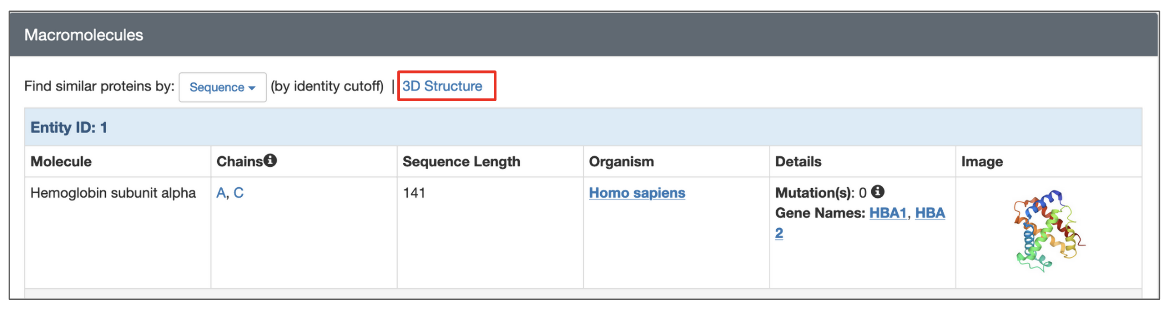
|
| Options to launch a structure based search from the structure summary page (highlighted in a red box). |
To search for assemblies similar to a specific assembly of the structure click on the Find Similar Assemblies link written below the snapshot of the assembly on the page.
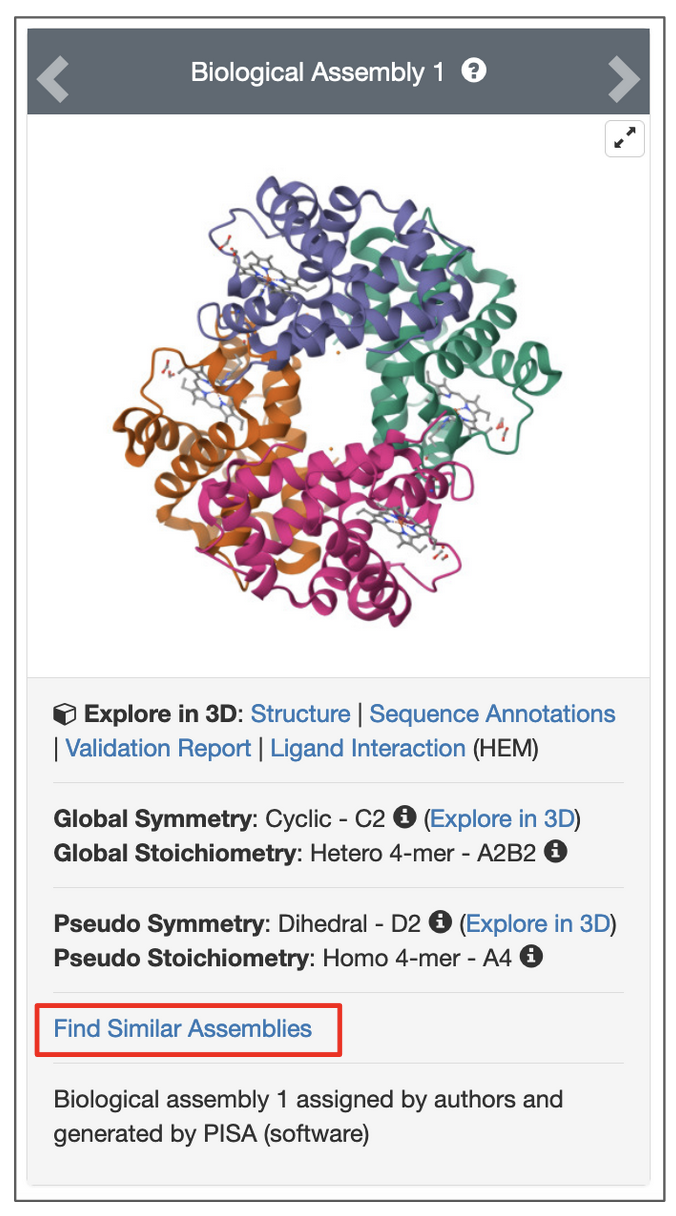
|
| Options to launch a search for an assembly from the structure summary page. Click on link highlighted in the red box. |
Search Results
Depending on the selected search options, the structure similarity results will list similar Macromolecules (Polymer Entities) or Assemblies.
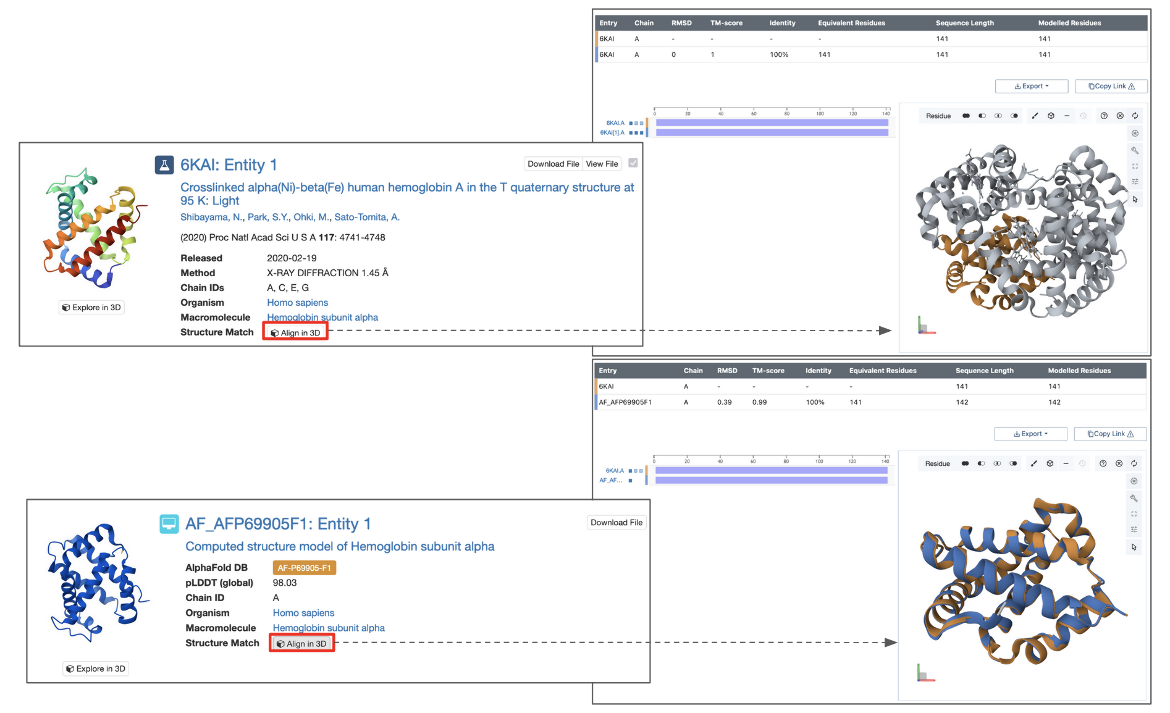
For chain-based searches, each search results can be superposed on the query chain and viewed interactively in 3D using the Pairwise Structure Alignment tool. Simply click the Align in 3D button next to “Structure Match” to open the alignment.
Note: This button is available only when the search is based on an existing PDB or CSM structure. It does not appear when the query structure is uploaded from a local file or provided via a URL.
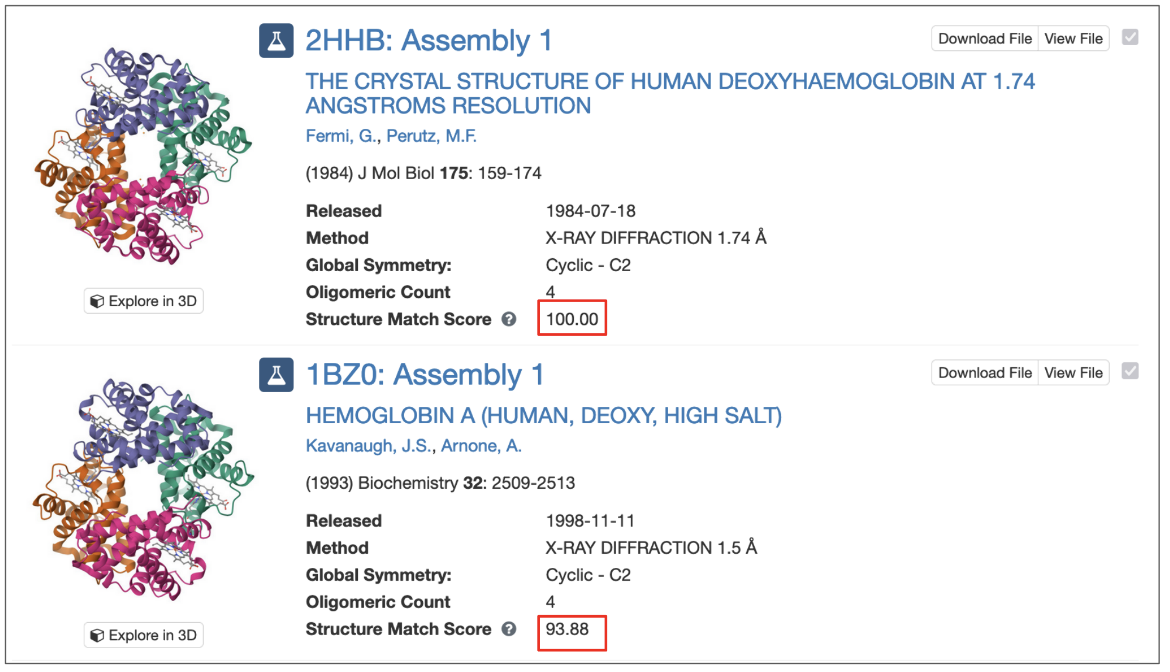
For assembly-based searches, each matched assembly is assigned a structure match score, which represents the probability (expressed as a percentage) that the assembly matches the query structure. A score of 100 indicates a perfect match, while lower values reflect decreasing levels of structural similarity.
Limitations of 3D Similarity Search
The structure similarity search system has some limitations:
- The method can not report an RMSD since it only produces a global optimal superposition of the volumes but knows nothing about residues that are paired in the alignment. Instead the method outputs a score that indicates the likelihood that the match is relevant.
- Highly symmetric assemblies often produce false positives (with lower scores), e.g. searching for a D3 point-group symmetric assembly will likely match a few unrelated D3 assemblies with lower scores.
- Flexible NMR structures will often be unmatched due to the long flexible tails
- Long protruding tails will result in failure to match otherwise globally similar shapes.
- The matching is global, thus local similarities are not found. For example:
- when searching for chains: 2 chains that are similar only in some common domain will usually not match,
- when searching for assemblies: 2 assemblies that are similar in some subset of chains but not globally will usually not match.
Examples
Find chains similar to Myoglobin
- Use the PDB/CSM ID option to select a chain from an existing PDB structure, such as pdb_00001mbn
- Chain A is selected by default
- Click the Search button to return matching polymer entities from the PDB archive
- Enable Include CSM and run the search again to retrieve both matching experimental structures and predicted models
Find chains similar to the open form of hexokinase
- Use a structure of the enzyme hexokinase in an “open” conformation as a query (PDB ID pdb_00002yhx, chain A)
- Click the Search button to return matching polymer entities
Find assemblies similar to the SARS-CoV-2 Spike protein trimer
- The SARS-CoV-2 spike protein is composed of three polymer chains, each of which has a receptor-binding domain that can be in an open (or up) conformation for interacting with cellular receptors or a closed (or down) conformation.
- To find spike structures where all three receptor-binding domains are closed, use PDB ID pdb_00006vxx as a query
- Change query structure selection to Assembly, Assembly 1 will be selected by default
- Click the Search button to return matching assemblies
Find assemblies similar to Insulin hexamers
- Use PDB ID pdb_00001trz as a query
- Change query structure selection to Assembly and change the query to match to Assembly 3
- Click the Search button to return matching assemblies
Find Insulin chains with a shape similar to mature Insulin (composed of two polymer chains)
- Use PDB ID pdb_00001trz as a query
- Change query structure selection to Assembly, Assembly 1 will be selected by default
- Change Search Targets option to match Chains
- Click the Search button to return matching polymer entities
Find assemblies similar to the Chymotrypsin polymer
- Use PDB ID pdb_00001k2i as a query
- Chain A [auth 1] is selected by default
- Change Search Targets option to match Assemblies
- Click the Search button to return matching assemblies
References
- Guzenko, D., Burley, S. K., Duarte, J. M. (2020) Real time structural search of the Protein Data Bank". PLoS Computational Biology, https://doi.org/10.1371/journal.pcbi.1007970
- Novotni, M., & Klein, R. (2004). Shape retrieval using 3D Zernike descriptors. Comput. Aided Des., 36, 1047-1062, https://doi.org/10.1016/j.cad.2004.01.005